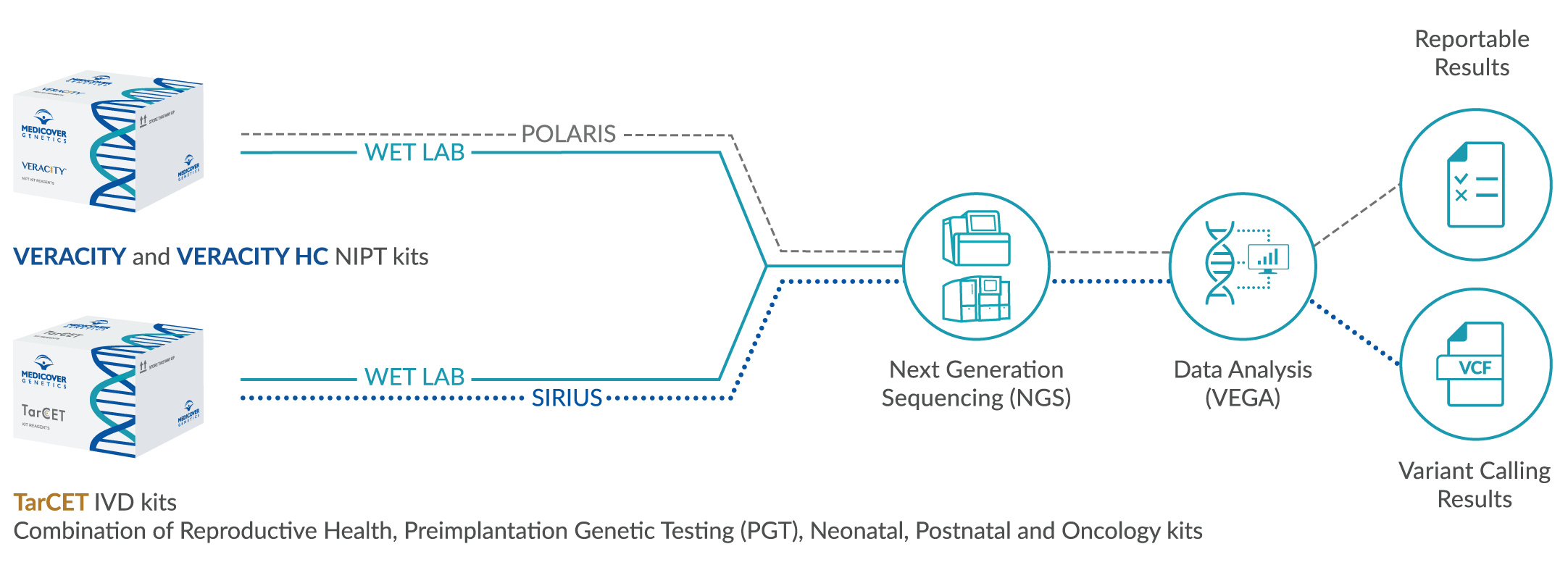
Antimicrobial resistance (AMR) is one of the most pressing global health threats, and accurate identification and surveillance of multidrug-resistant organisms (MDRO) are key to controlling its spread. A recent study shows that long-read sequencing offers an effective, cost-efficient alternative to traditional methods for tracking and identifying resistant pathogens. By comparing long-read and short-read sequencing, the study confirms that long-read sequencing delivers highly accurate genomic data, enabling precise surveillance and outbreak analysis of MDROs. This approach holds significant promise, especially for resource-limited settings, where it could enhance global efforts to fight AMR. With its potential for rapid, accurate, and low-cost genomic surveillance, this method is poised to support critical public health initiatives worldwide. Read more about this under Article 1 below.
Contents
- Article 1: Genomic surveillance of multidrug-resistant organisms based on long-read sequencing
- Article 2: Genome-wide association analysis provides insights into the molecular etiology of dilated cardiomyopathy
- Article 3: Curation and reporting of pathogenic genome-wide copy-number variants in a prenatal cell-free DNA screen
- Article 4: Management of individuals with heterozygous germline pathogenic variants in ATM: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG)
- Article 5: Unveiling genetic insights: Array-CGH and WES discoveries in a cohort of 122 children with essential autism spectrum disorder
- References
Article 1: Genomic surveillance of multidrug-resistant organisms based on long-read sequencing
This study showed that long-read sequencing accurately detects antimicrobial resistance genes and identifies outbreaks in multidrug-resistant organisms (MDROs), providing rapid and cost-effective solutions. It holds potential for improving global public health efforts against antimicrobial resistance, even in resource-limited settings. Read the full article here.
In summary: Long-read sequencing for multidrug-resistant organism surveillance
Article 2: Genome-wide association analysis provides insights into the molecular etiology of dilated cardiomyopathy
A genome-wide study identified 80 genetic risk loci and 62 potential effector genes for dilated cardiomyopathy (DCM). It uncovered cellular pathways involved in DCM pathogenesis and demonstrated that polygenic scores predict DCM risk, guiding genetic testing and therapy development. Read the full article here.
In summary: Identified genetic risk loci for dilated cardiomyopathy
Article 3: Curation and reporting of pathogenic genome-wide copy-number variants in a prenatal cell-free DNA screen
This study developed a framework for interpreting fetal copy-number variants (CNVs) in prenatal screening, improving accuracy and clarity in reporting. Applied to over 300,000 samples, it identified 65 syndromes, emphasizing a patient-centered approach for reproductive decision-making. Read the full article here.
In summary: New framework for fetal CNV reporting in prenatal screening
Article 4: Management of individuals with heterozygous germline pathogenic variants in ATM: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG)
An international workgroup provided guidance for managing carriers of ATM germline pathogenic variants, associated with increased cancer risks. Recommendations include enhanced surveillance for breast, prostate, and pancreatic cancers, with an emphasis on personalized risk and clinical trials. Read the full article here.
In summary: Guidelines for managing ATM germline pathogenic variant carriers
Article 5: Unveiling genetic insights: Array-CGH and WES discoveries in a cohort of 122 children with essential autism spectrum disorder
Whole exome sequencing (WES) and array-CGH identified pathogenic variants in 31.2% of children with essential autism spectrum disorder (ASD). The study found 138 new candidate genes, enhancing ASD genetic understanding and suggesting improved diagnostic and treatment strategies. Read the full article here.
In summary: WES and array-CGH improve genetic understanding of essential Autism Spectrum Disorder
References
[1] Landman, F., Jamin, C., de Haan, A., Witteveen, S., Bos, J., van der Heide, H. G. J., Schouls, L. M., Hendrickx, A. P. A., & Dutch CPE/MRSA surveillance study group (2024). Genomic surveillance of multidrug-resistant organisms based on long-read sequencing. Genome medicine, 16(1), 137. https://doi.org/10.1186/s13073-024-01412-6
[2] Zheng, S. L., Henry, A., Cannie, D., Lee, M., Miller, D., McGurk, K. A., Bond, I., Xu, X., Issa, H., Francis, C., De Marvao, A., Theotokis, P. I., Buchan, R. J., Speed, D., Abner, E., Adams, L., Aragam, K. G., Ärnlöv, J., Raja, A. A., Backman, J. D., … Lumbers, R. T. (2024). Genome-wide association analysis provides insights into the molecular etiology of dilated cardiomyopathy. Nature genetics, 56(12), 2646–2658. https://doi.org/10.1038/s41588-024-01952-y
[3] Cox, S. G., Acevedo, A., Ahuja, A., LaBreche, H. G., Alfaro, M. P., Pierson, S., Westover, T., Ratzel, S., Hancock, S., Moyer, K., & Muzzey, D. (2024). Curation and reporting of pathogenic genome-wide copy-number variants in a prenatal cell-free DNA screen. Genetics in medicine : official journal of the American College of Medical Genetics, 101223. Advance online publication. https://doi.org/10.1016/j.gim.2024.101223
[4] Pal, T., Schon, K. R., Astiazaran-Symonds, E., Balmaña, J., Foulkes, W. D., James, P., Klugman, S., Livinski, A. A., Mak, J. S., Ngeow, J., Voian, N., Wick, M. J., Hanson, H., Stewart, D. R., Tischkowitz, M., & ACMG Professional Practice and Guidelines Committee. Electronic address: documents@acmg.net (2024). Management of individuals with heterozygous germline pathogenic variants in ATM: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genetics in medicine : official journal of the American College of Medical Genetics, 101243. Advance online publication. https://doi.org/10.1016/j.gim.2024.101243
[5] Granata, P., Zito, A., Cocciadiferro, D., Novelli, A., Pessina, C., Mazza, T., Ferri, M., Piccinelli, P., Luoni, C., Termine, C., Fasano, M., & Casalone, R. (2024). Unveiling genetic insights: Array-CGH and WES discoveries in a cohort of 122 children with essential autism spectrum disorder. BMC genomics, 25(1), 1186. https://doi.org/10.1186/s12864-024-11077-5