OVERVIEW
Hereditary hearing loss has a prevalence of 1 in 500 newborns and are one of the most common congenital disorders. In adults, the prevalence is increasing to 3.5:1,000. The proportion of genetically caused deafness is about 50-70%. Inherited hearing loss disorders can be syndromic or non-syndromic. Syndromic hearing disorders are associated with malformations of the external ear with involvement of other organs. Non-syndromic hearing disorders are associated with abnormalities of the middle ear and/or inner ear but do not have visible abnormalities of the external ear or involvement of any other medical problem. More than 70% of genetic deafness is non-syndromic, about 80% of non-syndromic genetic deafness follows an autosomal recessive inheritance. With the investigation of the genes GJB2 and GJB6 about 50% of the cases with autosomal recessive, non-syndromic, sensorineural deafness can be clarified. Early identification of individuals at risk can help establish the right clinical management plan.
We offer comprehensive and syndrome-specific panels testing for hearing loss disorders. The test can offer a molecular genetic diagnosis of a hearing loss disorder that is observed or predicted in you or a family member.

IMPORTANCE OF GETTING TESTED
If you or a family member has a risk of a hearing loss disorder, identifying the cause can help to take actions to improve the outcome of the disorder. Additionally, family members can be informed and encouraged to also get tested. Our genetic counsellors can provide medical advice.

You have a child with
delayed speech development

You have a young child with
autism spectrum disorder
or specific language disorders

You want to estimate
the risk of recurrence

POSSIBLE OUTCOMES OF THE TEST
A molecular genetic diagnostic report outlining the results of the sequencing analysis is provided. Changes in DNA sequences (variants) can be detrimental and lead to the development of a cardiac or aortic disorder, including asymptomatic disorders that develop later in life. We will report pathogenic and likely pathogenic variants as well as variants of unknown significance.
Pathogenic and likely pathogenic variants mean the genetic cause of the observed symptoms has been identified and may help determine the right treatment and management plan.
Variants of unknown significance means there was not enough evidence to classify the variant as either pathogenic or neutral. Annual variant reclassification and testing family members is recommended.
It is important to note that a negative result does not guarantee the absence of a disorder or that the disorder does not have a genetic cause. Genetic testing is an evolving field and may not detect all variants or there may not currently be enough evidence to classify all variants that lead to an inherited disease.
MEDICAL GENETIC COUNSELLING
We provide expert medical genetic counselling as part of a genetic testing journey. Genetic counselling is a process of communication that supports patients and their relatives before and after genetic testing. It is educational, impartial and nondirective. Prior to any genetic test, genetic counsellors will obtain a detailed family history, explain the method of testing that will be used, its risks and benefits, the limitations of the diagnosis and the implications of making a genetic diagnosis (Elliott and Friedman, 2018, Nat Rev Genet 19:735).
Upon receiving the genetic test results, genetic counselling can help the specialist physician and the patient to interpret them. They can be advised of the consequences of the results including the probability of developing the genetic disorder or passing it on to children, as well as ways to prevent, avoid or reduce these risks (Yang and Kim, 2018, Ann Lab Med 38:291). Our goal of counselling is to provide the patient with greater knowledge and thus, a better understanding of the results and the ability to make a more informed decision.

1 ml EDTA Blood
Please note: DO NOT FREEZE. Ship with a frozen ice pack. Prepare the package such that the tube is tightly packed and not loose. Avoid getting the blood tube wet from the ice pack. Place a paper towel in between ice pack and blood tube if necessary. In sub-zero temperatures, include an unfrozen ice pack in the shipping container as insulation.
15-25 working days
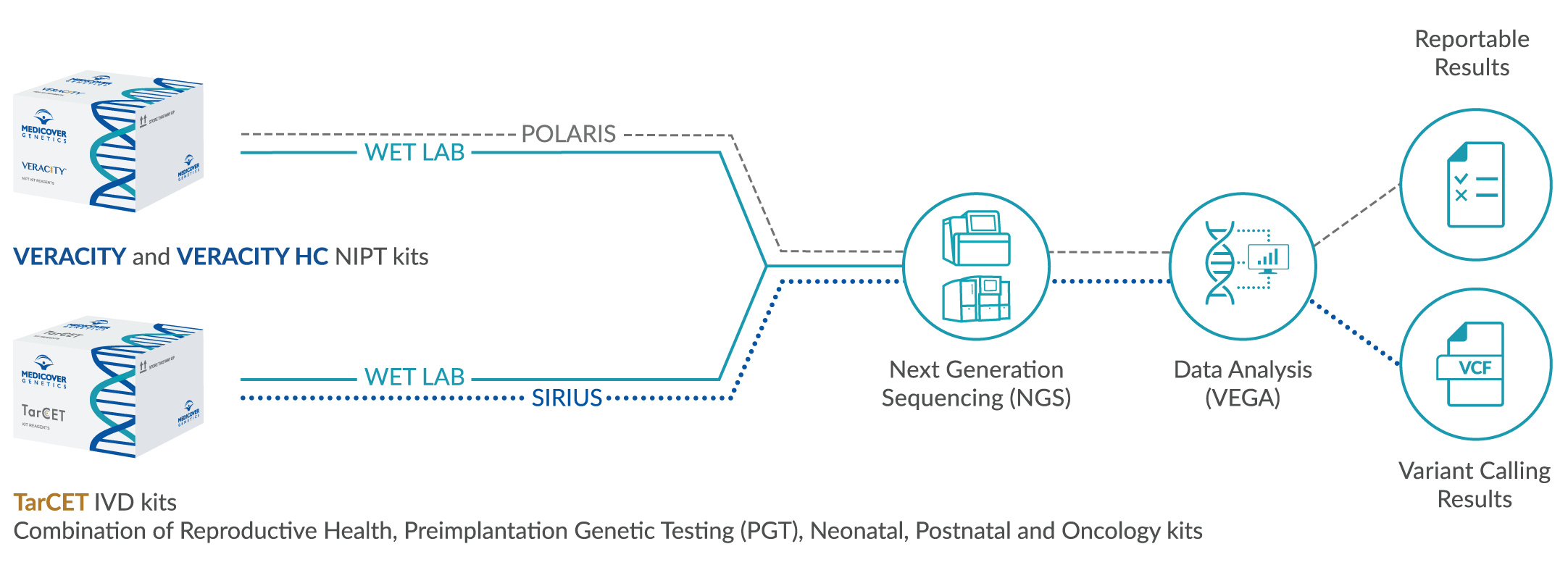
DNA is isolated and next generation sequencing is performed on all coding exons and conserved intronic regions. Single base pair changes, small deletions and duplications and copy number variants (CNV) are identified. Sequencing runs result in a Quality Score of >30 (accuracy >99.9%) in at least 75% of all bases with a coverage of >20-fold. CNV detection sensitivity is 76.99% and precision is 62.59% (with GC limitation between 0.4 and 0.6 per target sensitivity is 77.04% and precision is 84.10%). Variant classification is performed following ACMG guidelines (Richards et al. 2015, Genet Med 17:405; Kearney et al. 2011, Genet Med 13:680).
Next generation
sequencing (Illumina)
Twist Human Core
Exome plus Ref Seq
Spikeln
Illumina DRAGEN
Bio-IT Platform
VarSeq by
GoldenHelix
hg38, NCBI GR38
>30 (precision >99,9%)
in min. 75% of bases
99.92-99.93%; confirmation of reported SNV with Sanger
sequencing, data analysis with
SeqPilot
Richards et al. 2015, Genet Med
17:405; Ellard et al. “ACGS Best
Practice Guidelines for Variant
Classification 2020″
MaxEntScan,
SpliceSiteFinder-like,
REVEL
HGMD Professional
release, ClinVar,
gnomAD
OUR TESTS
Genes: ABHD12, ADGRV1, AIFM1, ATP6V1B1, BSND, CACNA1D, CDH23, CLPP, CLRN1, DIAPH3, EDN3, EDNRB, ERAL1, EYA1, FOXI1, GPSM2, HARS, HSD17B4, KCNE1, KCNJ10, KCNQ1, LARS2, MITF, MYH9, MYO7A, PAX3, PCDH15, PDZD7, SIX5, SLC26A4, SLITRK6, SNAI2, SOX10, TWNK, USH1C, USH1G, USH2A, WFS1, WHRN
Genes: GJB2, GJB6
Genes: ABHD12, ACTG1, ADCY1, ADGRV1, AIFM1, ATP6V1B1, BDP1, BSND, CABP2, CACNA1D, CCDC50, CD164, CDC14A, CDH23, CEACAM16, CEP250, CIB2, CLDN14, CLDN9, CLIC5, CLPP, CLRN1, COCH, COL11A1, COL11A2, COL4A6, CRYM, DCDC2, DIABLO, DIAPH1, DIAPH3, DMXL2, EDN3, EDNRB, ELMOD3, EPS8, EPS8L2, ERAL1, ESPN, ESRP1, ESRRB, EYA1, EYA4, FAM189A2, FOXI1, GAB1, GIPC3, GJB2, GJB3, GJB6, GPSM2, GRAP, GRHL2, GRXCR1, GRXCR2, GSDME, HARS, HARS2, HGF, HOMER2, HSD17B4, ILDR1, KARS, KCNE1, KCNJ10, KCNQ1, KCNQ4, KITLG, LARS2, LHFPL5, LMX1A, LOXHD1, LRTOMT, MARVELD2, MCM2, MET, MIR182, MIR183, MIR96, MITF, MPZL2, MSRB3, MYH14, MYH9, MYO15A, MYO3A, MYO6, MYO7A, NARS2, NLRP3, OSBPL2, OTOA, OTOF, OTOG, OTOGL, P2RX2, PAX3, PCDH15, PDE1C, PDZD7, PJVK, PLS1, PNPT1, POU3F4, POU4F3, PPIP5K2, PRPS1, PTPRQ, RDX, REST, RIPOR2, ROR1, S1PR2, SERPINB6, SIX1, SIX5, SLC17A8, SLC22A4, SLC26A4, SLC26A5, SLITRK6, SMPX, SNAI2, SOX10, SPNS2, STRC, SYNE4, TBC1D24, TECTA, TJP2, TMC1, TMEM132E, TMIE, TMPRSS3, TNC, TPRN, TRIOBP, TSPEAR, TWNK, USH1C, USH1G, USH2A, WBP2, WFS1, WHRN
Genes: SLC26A4
Genes: CLPP, ERAL1, HARS2, HSD17B4, LARS2, TWNK
Genes: ADGRV1, CDH23, CIB2, CLRN1, HARS, MYO7A, PCDH15, PDZD7, USH1C, USH1G, USH2A, WHRN
Genes: EDN3, EDNRB, MITF, PAX3, SNAI2, SOX10
Genes: WFS1
LATEST ARTICLES