OVERVIEW
WHAT IS NEOTHETIS?NeoThetis is a non-invasive genetic test that can guide therapy selection for people diagnosed with cancer via a simple blood draw. During cancer development and growth, cancer cells release DNA into the bloodstream known as circulating tumor DNA (ctDNA). ctDNA contains genetic characteristics of the tumor which contribute to cancer development and therapy resistance.
NeoThetis is a novel liquid biopsy test that accurately detects and analyses the ctDNA released from both primary and metastatic tumors of patients diagnosed with cancer. It is specifically designed to maximize the available therapeutic opportunities which are based on precision medicine while guiding therapy selection and treatment re-evaluation tailored to each patient.
● Performed non-invasively, using a simple blood draw
● Guides therapy selection, and treatment re-evaluation for therapy resistance.
● Provides therapy solutions approved by professional cancer societies and bodies.
● Suggests available ongoing clinical trials.
● Identifies potential mutations that might arise in distant non-operable metastatic lesions.
● Captures the genetic characteristics of the tumor offering a deep investigation of the genetic alterations and immunotherapy biomarkers.
● Offers fast turnaround results, ensuring treatment can begin faster.


WHEN TO PERFORM NEOTHETIS?
NeoThetis can be performed at initial diagnosis and during disease progression:
● when tumor is inaccessible
● when tissue biopsy is limited or not available
● when the patient is unfit for tumor biopsy
● if additional tumor biopsy procedures are not clinically recommended
● when fast turnaround time is required for therapy selection
● when the patient does not respond to current treatment and re-evaluation is needed
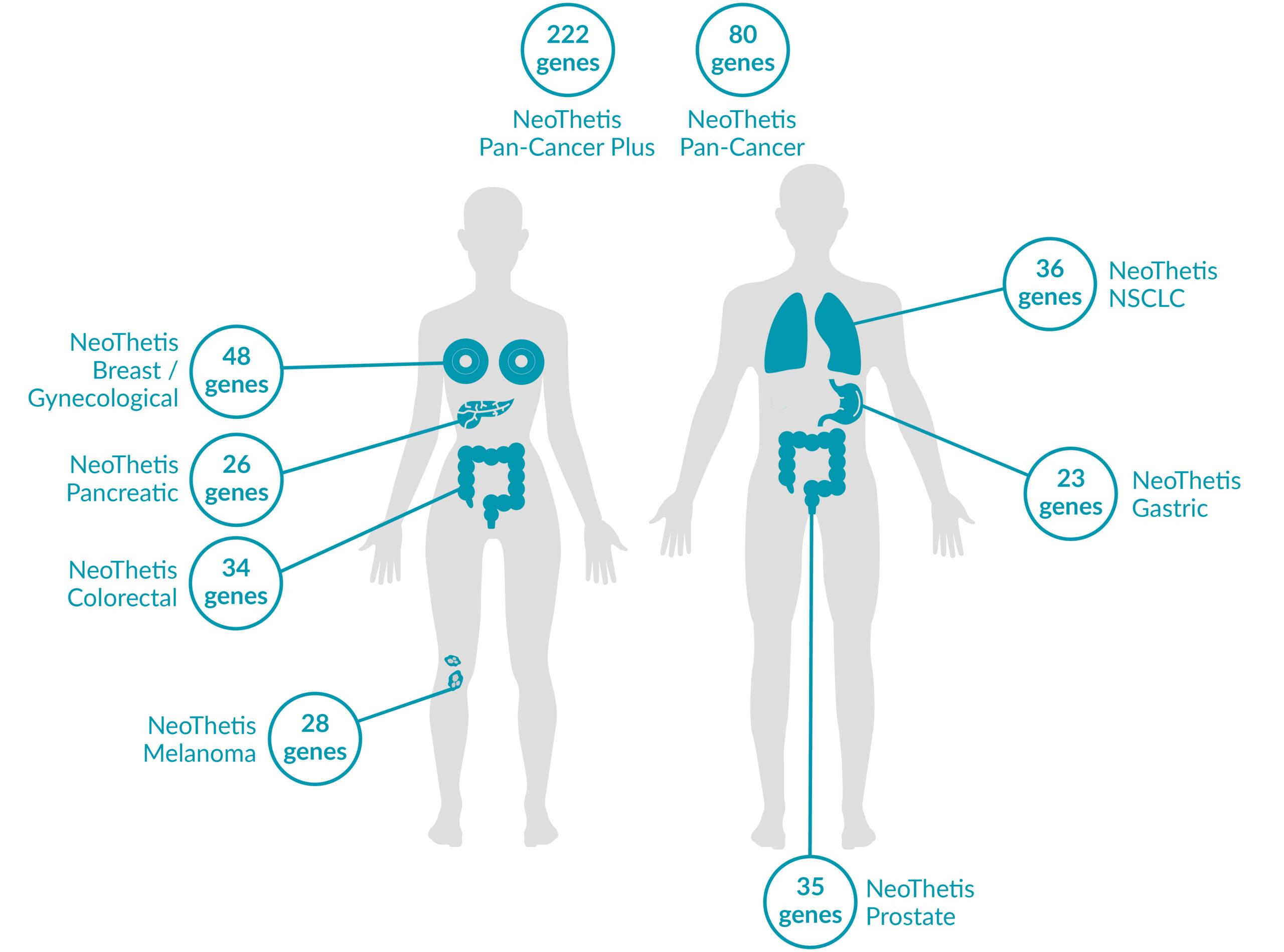
OUR TESTS
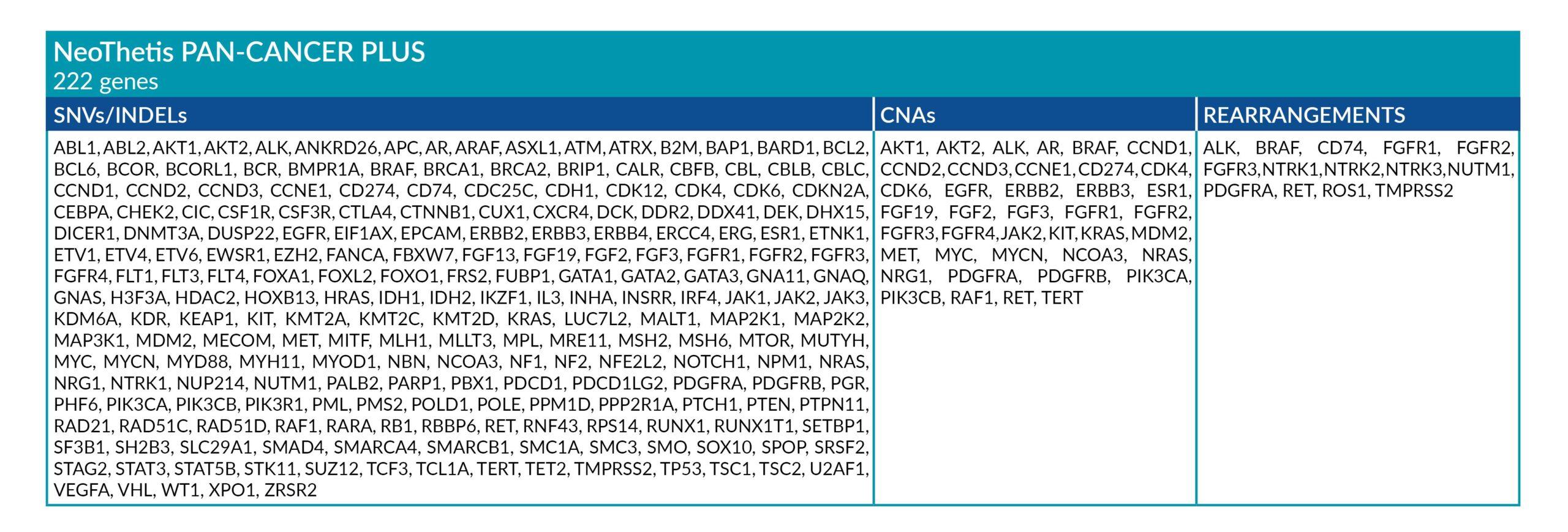
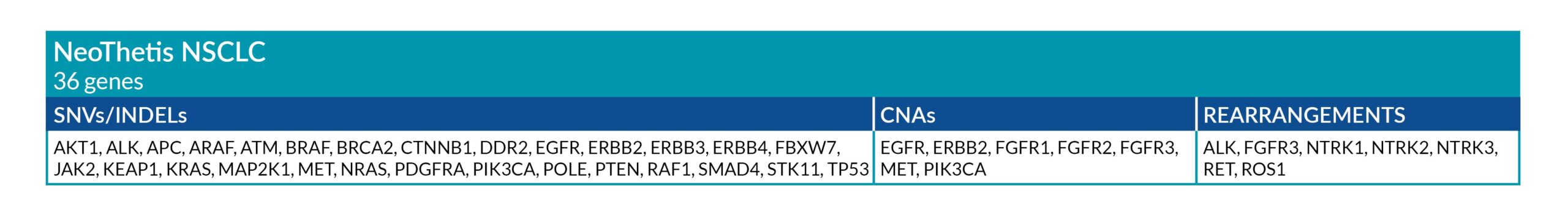
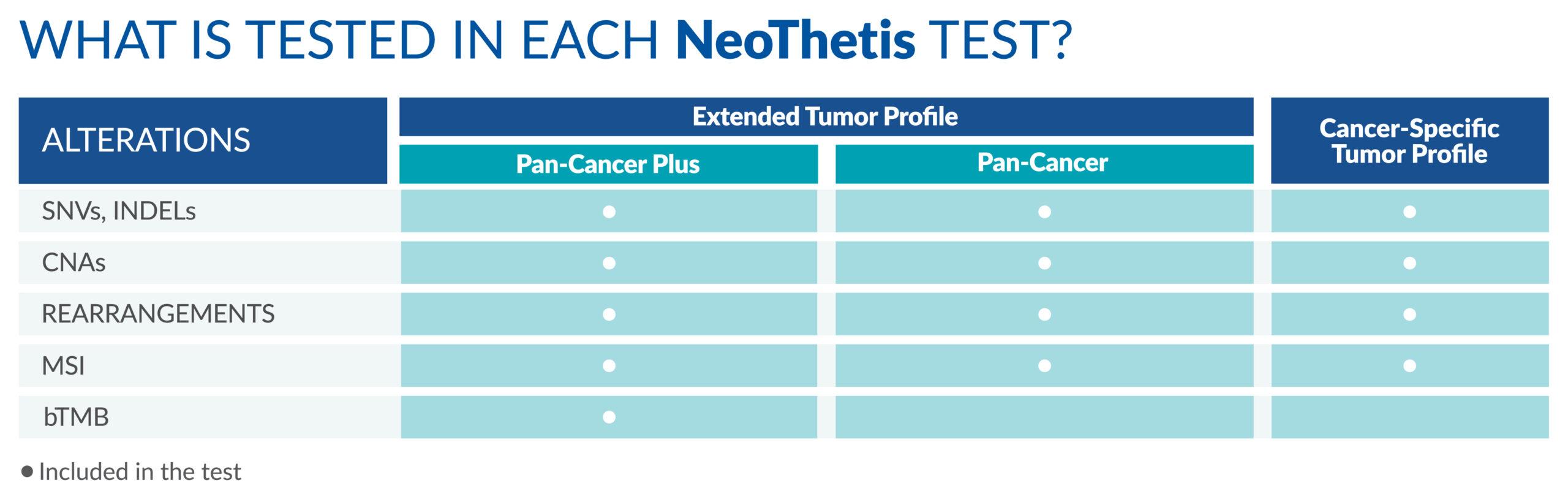
NeoThetis tests are designed to detect clinically actionable genetic alterations, including Single Nucleotide Variants (SNVs), Insertions and Deletions (INDELs), Copy Number Amplifications (CNAs), and Rearrangements, that drive cancer or are associated with response to treatment.
All NeoThetis tests also assess Microsatellite Instability (MSI) immunotherapy biomarker, with the Pan-Cancer Plus test additionally testing for blood Tumor Mutational Burden (bTMB) immunotherapy biomarker.
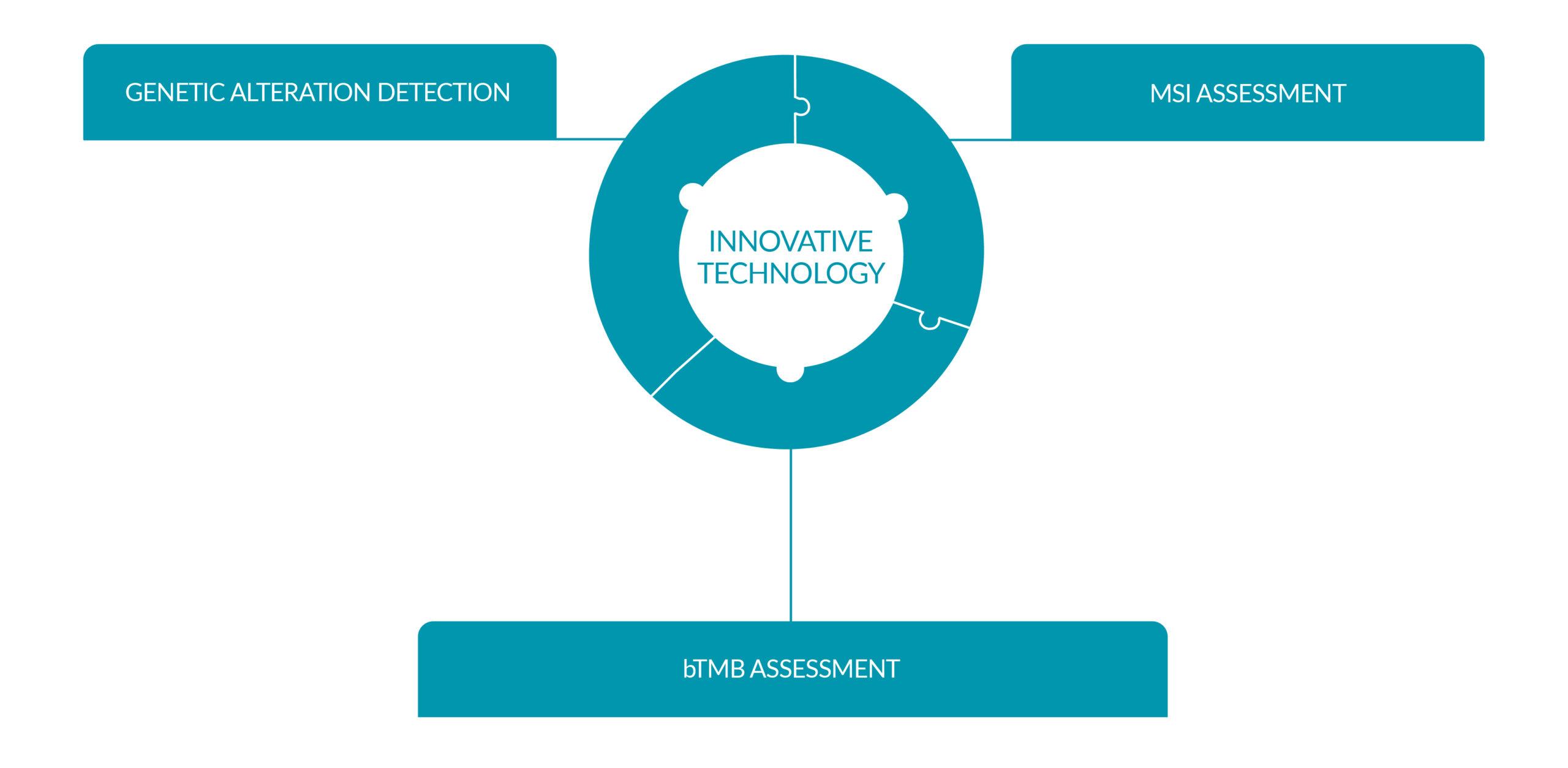
INNOVATIVE TECHNOLOGICAL DESIGN
Targeted technology and novel bioinformatics
Unparalleled workflow which combines our proprietary targeted capture enrichment technology along with novel bioinformatic pipelines, to provide accurate detection of genetic variants even at low allele frequencies. Our high read depth analysis enables for increased sensitivity and specificity providing reliable results.
Genetic alteration detection
NeoThetis screens clinically important coding regions and selected non-coding regions in genes of interest, to identify genetic alterations which:
● are included in NCCN, ESMO and ASCO guidelines
● are associated with FDA/EMA approved therapies
● serve as selection criteria in clinical trials
● are associated with therapy resistance
MSI assessment
MSI is an immunotherapy biomarker caused by defects in the mismatch repair mechanism.
MSI testing via Next Generation Sequencing (NGS) detects a higher number of clinically significant loci compared to other MSI testing methods, such as immunohistochemistry, with high accuracy and sensitivity. MSI assessment:
● has been emphasized by cancer societies including NCCN, ESMO and ASCO
● is associated with FDA/EMA approved immunotherapy drugs
bTMB assessment
bTMB is an emerging immunotherapy which indicates the total number of mutations found in a tumor per megabase and it is obtained from analyzing ctDNA. bTMB is tumor agnostic and can guide healthcare providers to identify patients who might benefit from immunotherapy.
GENETIC ALTERATIONS AND BIOMARKERS TESTED
RECOMMENDATIONS OF LIQUID BIOPSY BY PROFESSIONAL BODIES AND SOCIETIES
The use of cell-free/circulating tumor DNA testing can be considered when a patient is medically unfit for invasive tissue sampling, or if in the initial diagnostic setting there is insufficient material for molecular analysis following pathologic confirmation.
“Validated and sensitive ctDNA assays can be used to genotype advanced cancers and select patients for targeted therapies”


POSSIBLE OUTCOMES OF THE TEST
A detailed report outlining the results of the genetic test is provided:
● IMMUNOTHERAPY BIOMARKERS
MSI status and bTMB score is reported, accompanied with available FDA/EMA approved drugs which are eligible for the variants detected.
● GENOMIC FINDINGS
All the genetic variants identified are reported. The results will also indicate which approved FDA/EMA targeted therapies are suitable as treatment options. Depending on the results identified, the report will also provide information on the drugs the patient will not benefit from.
Variants of strong and of potential clinical significance as well as variants of uncertain significance (VUS) are also reported.
● CLINICAL TRIALS
Provides information on clinical trials which are relevant to the genetic variants identified.
● INTERPRETATION
The biological and therapeutical classification of results are reported in detail according to NCCN guidelines