UPDATES IN PROGRESS
We’re currently working on updating this section. Please check back soon!
In the meantime, you can visit our German Pharmacogenetics webpage for more information.






This browser is not supported, please use a modern browser!

We’re currently working on updating this section. Please check back soon!
In the meantime, you can visit our German Pharmacogenetics webpage for more information.


OUR NETWORK
Medicover Genetics’ technology transfer platform offers comprehensive testing in a single sequencing run, making it a unique and efficient solution for laboratories of any size. With our Technology Transfer platform, laboratories can perform multi-disciplinary genetic tests such as NIPT, cardiac, hereditary cancer, carrier screening, and metabolic testing all in a single sequencing run. This improves turnaround time and reduces operational costs while ensuring high-quality results with no compromise on accuracy. Our comprehensive testing approach is ideal for laboratories looking to provide a wide range of genetic tests to their healthcare associates and patients.
Our Technology Transfer platform workflow is designed to minimize the risk of errors, reduce hands-on processing and increase the accuracy of test results, providing confidence to patients and physicians in making informed decisions with our user-friendly protocols. The implementation of these protocols ensures that laboratory personnel are trained to follow standardized procedures, reducing the likelihood of human error, and increasing the quality of results.
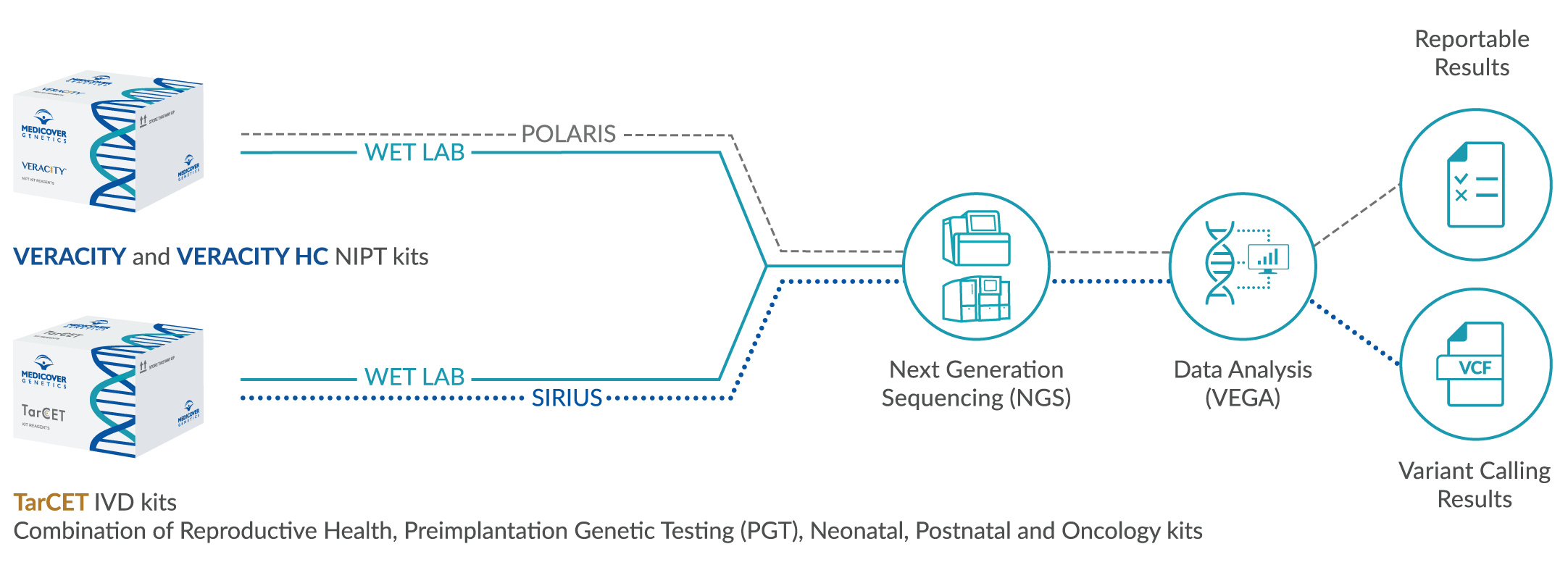
VEGA is a bioinformatics analysis software for samples that have been processed using NIPT IVD kits — VERACITY and VERACITY High Content (HC) — or TarCET kits. VEGA performs analysis on NGS data generated by the NIPT or TarCET workflows. The NIPT analysis provides fetal fraction estimation and binary classification of samples into high risk or low risk for trisomy 21, 18, 13, Sex Chromosome Aneuploidies (SCAs), selected microdeletions, and monogenic diseases. The TarCET analysis detects single nucleotide variants, short insertions and/or deletions and copy number alterations. Additionally, VEGA can be used for TarCET PGT to detect aneuploidies and structural rearrangements in embryos created for In Vitro Fertilisation (IVF).
Medicover Genetics’ versatile Technology Transfer platform can be integrated in a single workflow, offering a multidisciplinary portfolio of genetic tests. VERACITY, VERACITY High Content (HC) and TarCET kits are CE-marked and include user friendly color identification formatting. VERACITY and VERACITY HC kits contain 96 reactions and TarCET kits contain 16 reactions.



Our Technology Transfer workflow integrates VERACITY and VERACITY High Content (HC) NIPT and TarCET kits for the provision of in-house genetic testing. Multiple assays can be run simultaneously in consolidated workflows, with minimized hands-on time and re-runs, maintaining laboratory proficiency and sample handling efficiency.
Our Technology Transfer workflow delivers accurate results, while the pre- and post-deployment process initiated by Medicover Genetics, is focused on delivering excellent customer service, efficient operational management, laboratory and commercial training, and solutions for unique laboratory pain points.
Description
FH, PH and RAS Panel analyzes 11, 11 and 30 genes and covers Familial Hypercholesterolemia (FH), Pulmonary Hypertension (PH) and RASopathy (RAS) related disorders, respectively.
Genes Tested for FH
ABCA1, ABCG5, ABCG8, APOA5, APOB, APOE, LDLR, LDLRAP1, LIPA, LPL, PCSK9
Genes Tested for PH
ACVRL1, BMPR1B, BMPR2, CAV1, EIF2AK4, ENG, KCNA5, KCNK3, SMAD4, SMAD9, TBX4
Genes Tested for RASopathy
AKT3, BRAF, CBL, CCND2, EPHB4, HRAS, KRAS, LZTR1, MAP2K1, MAP2K2, MRAS, NF1, NF2, NRAS, PIK3CA, PIK3R2, PPP1CB, PTPN11, RAF1, RASA1, RASA2, RIT1, RRAS, SASH1, SHOC2, SMARCB1, SOS1, SOS2, SPRED1, STAMBP
Technical Specifications
● Genomic regions not covered by FH:
• NM_000384.3 (APOB): exon 1
• NM_015627.3 (LDLRAP1): exon 1
• NM_000237.3 (LPL): exon 5
● Genomic regions not covered by PH:
• Not Applicable
● Genomic regions not covered by RAS:
• NM_001374258.1 (BRAF): exon 10 and exon 20
• NM_001042492.3 (NF1): exon 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,28, 29, 32, 33, 34, 35 and 36
• NM_005027.4 (PIK3R2): exon 6
Technical Limitations for FH, PH and RAS
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Congenital Heart Defects Panel analyzes 80 genes and covers inherited congenital heart defects.
Genes Tested
ACTA2, ACTC1, ACVR2B, AFF4, BMPR2, CAD, CFAP53, CDK13, CELSR1, CELSR2, CELSR3, CHD4, CHD7, CITED2, CREBBP, CRELD1, DNAH11, DNAH5, DNAH6, DNAI1, DTNA, EHMT1, ELN, EVC, EVC2, FBN1, FLNA, FOXC1, FOXH1, GANAB, GATA4, GATA5, GATA6, GDF1, GPC3, HAND1, HAND2, HRAS, JAG1, KDM5B, KMT2D, MED13L, MMP21, MYH11, MYH6, MYH7, NIPBL, NKX2-5, NKX2-6, NME7, NODAL, NOTCH1, NOTCH2, NR2F2, PITX2, PKD1L1, PLD1, POGZ, PRDM6, PRKD1, RABGAP1L, RBFOX2, RBM10, SALL4, SEMA3D, SEMA3E, SMAD6, TAB2, TBX1, TBX20, TBX5, TFAP2B, TGFBR1, TGFBR2, TLL1, TMEM260, TPM1, ZEB2, ZFPM2, ZIC3
Technical Specifications
● Genomic regions not covered by this test:
• NM_005159.5 (ACTC1): exon 2
• NM_001106.4 (ACVR2B): exon 1
• NM_024757.5 (EHMT1): exon 1
• NM_153717.3 (EVC): exon 1
• NM_001110556.2 (FLNA): exon 15
• NM_001308093.3 (GATA4): exon 1
• NM_001492.6 (GDF1): exon 8
• NM_000214.3 (JAG1): exon 1
• NM_017617.5 (NOTCH1): exon 1
• NM_001136239.4 (PRDM6): exon 2
• NM_001349999.2 (RBFOX2): exon 1
• NM_005585.5 (SMAD6): exon 1
• NM_080647.1 (TBX1): exon 8, and 9
• NM_004612.4 (TGFBR1): exon 1
• NM_012082.4 (ZFPM2): exon 1
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Aortopathy Panel analyzes 48 genes and covers aortopathy-related inherited cardiovascular disorders.
Genes Tested
ABCC6, ACTA2, ACVR1, ADAMTS2, ALDH18A1, ATP6V0A2, ATP6V1A, ATP6V1E1, B3GALT6, B4GALT7, BGN, C1R, C1S, CBS, CHST14, COL1A1, COL1A2, COL3A1, COL4A1, COL5A1, COL5A2, DSE, EFEMP2, ELN, FBLN5, FBN1, FBN2, FKBP14, FOXE3, GORAB, LOX, LTBP4, MFAP5, MYH11, MYLK, PLOD1, PRDM5, PYCR1, SKI, SLC2A10, SLC39A13, SMAD3, SMAD4, TGFB2, TGFB3, TGFBR1, TGFBR2, ZNF469
Technical Specifications
● Genomic regions not covered by this test:
• NM_014244.5 (ADAMTS2): exon 1
• NM_007255.3 (B4GALT7): exon 1
• NM_000071.3 (CBS): exon 15
• NM_000089.4 (COL1A2): exon 3
• NM_001278074.1 (COL5A1): exon 1
• NM_001042545.2 (LTBP4): exon 4
• NM_004612.4 (TGFBR1): exon 1
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Arrhythmia Panel analyzes 42 genes and covers arrhythmia-related inherited cardiovascular disorders.
Genes Tested
ABCC9, AKAP9, ANK2, CACNA1C, CACNA2D1, CACNB2, CALM1, CALM2, CALM3, CASQ2, CAV3, GJA5, GNAI2, GPD1L, HCN4, KCNA5, KCNAB2, KCND3, KCNE1, KCNE2, KCNE3, KCNH2, KCNJ2, KCNJ5, KCNJ8, KCNQ1, LMNA, MYL4, NPPA, RYR2, SCN10A, SCN1B, SCN2B, SCN3B, SCN4B, SCN5A, SNTA1, TBX5, TECRL, TNNI3, TRDN, TRPM4
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Cardiomyopathy Panel analyzes 98 genes and covers cardiomyopathy-related inherited cardiovascular disorders.
Genes Tested
AARS2, ABCC9, ACAD9, ACADVL, ACTA1, ACTC1, ACTN2, AGK, AKAP9, ALMS1, ALPK3, ANKRD1, BAG3, CASQ2, CAV3, COA5, COA6, COX15, CPT2, CRYAB, CSRP3, CTNNA3, DES, DMD, DNAJC19, DOLK, DSC2, DSG2, DSP, DTNA, ELAC2, EMD, FHL1, FKTN, FLNC, GAA, GATAD1, GBE1, GLA, GTPBP3, HADHA, HADHB, HCN4, HRAS, ILK, JPH2, JUP, KARS1, LAMA4, LAMP2, LDB3, LMNA, MIB1, MRPL3, MRPL44, MTO1, MYBPC3, MYH6, MYH7, MYL2, MYL3, MYLK2, MYOZ2, MYPN, NDUFB11, NDUFV2, NEBL, NEXN, PDLIM3, PKP2, PLN, PPA2, PRDM16, PRKAG2, RAF1, RBM20, RYR2, SCN5A, SCO2, SGCD, SLC22A5, SLC25A20, SLC25A3, SLC25A4, TAFAZZIN, TCAP, TGFB3, TK2, TMEM43, TMEM70, TNNC1, TNNI3, TNNT2, TPM1, TSFM, TTN, TTR, VCL
Technical Specifications
● Genomic regions not covered by this test:
• NM_000018.4 (ACADVL): exon 2
• NM_001943.5 (DSG2): exon 1
• NM_001458.5 (FLNC): exon 20
• NM_021167.5 (GATAD1): exon 1
• NM_020433.5 (JPH2): exon 4
• NM_002230.4 (JUP): exon 11
• NM_016203.4 (PRKAG2): exon 13
• NM_014000.3 (VCL): exon 11
• NM_001267550.2 (TTN): exon 149, 164, 175, 176, 177, 184, 186, 193, 194, 195, 215
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Cardiac Comprehensive Panel analyzes 292 genes and covers major inherited cardiovascular disorders.
Genes Tested
AARS2, ABCA1, ABCC6, ABCC9, ABCG5, ABCG8, ACAD9, ACADVL, ACTA1, ACTA2, ACTC1, ACTN2, ACVR1, ACVR2B, ACVRL1, ADAMTS2, AFF4, AGK, AKAP9, AKT3, ALDH18A1, ALMS1, ALPK3, ANK2, ANKRD1, APOA5, APOB, APOE, ATP6V0A2, ATP6V1A, ATP6V1E1, B3GALT6, B4GALT7, BAG3, BGN, BMPR1B, BMPR2, BRAF, C1R, C1S, CACNA1C, CACNA2D1, CACNB2, CAD, CALM1, CALM2, CALM3, CASQ2, CAV1, CAV3, CBL, CBS, CFAP53, CCND2, CDK13, CELSR1, CELSR2, CELSR3, CHD4, CHD7, CHST14, CITED2, COA5, COA6, COL1A1, COL1A2, COL3A1, COL4A1, COL5A1, COL5A2, COX15, CPT2, CREBBP, CRELD1, CRYAB, CSRP3, CTNNA3, DES, DMD, DNAH11, DNAH5, DNAH6, DNAI1, DNAJC19, DOLK, DSC2, DSE, DSG2, DSP, DTNA, EFEMP2, EHMT1, EIF2AK4, ELAC2, ELN, EMD, ENG, EPHB4, EVC, EVC2, FBLN5, FBN1, FBN2, FHL1, FKBP14, FKTN, FLNA, FLNC, FOXC1, FOXE3, FOXH1, GAA, GANAB, GATA4, GATA5, GATA6, GATAD1, GBE1, GDF1, GJA5, GLA, GNAI2, GORAB, GPC3, GPD1L, GTPBP3, HADHA, HADHB, HAND1, HAND2, HCN4, HRAS, ILK, JAG1, JPH2, JUP, KARS1, KCNA5, KCNAB2, KCND3, KCNE1, KCNE2, KCNE3, KCNH2, KCNJ2, KCNJ5, KCNJ8, KCNK3, KCNQ1, KDM5B, KMT2D, KRAS, LAMA4, LAMP2, LDB3, LDLR, LDLRAP1, LIPA, LMNA, LOX, LPL, LTBP4, LZTR1, MAP2K1, MAP2K2, MED13L, MFAP5, MIB1, MMP21, MRAS, MRPL3, MRPL44, MTO1, MYBPC3, MYH11, MYH6, MYH7, MYL2, MYL3, MYL4, MYLK, MYLK2, MYOZ2, MYPN, NDUFB11, NDUFV2, NEBL, NEXN, NF1, NF2, NIPBL, NKX2-5, NKX2-6, NME7, NODAL, NOTCH1, NOTCH2, NPPA, NR2F2, NRAS, PCSK9, PDLIM3, PIK3CA, PIK3R2, PITX2, PKD1L1, PKP2, PLD1, PLN, PLOD1, POGZ, PPA2, PPP1CB, PRDM16, PRDM5, PRDM6, PRKAG2, PRKD1, PTPN11, PYCR1, RABGAP1L, RAF1, RASA1, RASA2, RBFOX2, RBM10, RBM20, RIT1, RRAS, RYR2, SALL4, SASH1, SCN10A, SCN1B, SCN2B, SCN3B, SCN4B, SCN5A, SCO2, SEMA3D, SEMA3E, SGCD, SHOC2, SKI, SLC22A5, SLC25A20, SLC25A3, SLC25A4, SLC2A10, SLC39A13, SMAD3, SMAD4, SMAD6, SMAD9, SMARCB1, SNTA1, SOS1, SOS2, SPRED1, STAMBP, TAB2, TAFAZZIN, TBX1, TBX20, TBX4, TBX5, TCAP, TECRL, TFAP2B, TGFB2, TGFB3, TGFBR1, TGFBR2, TK2, TLL1, TMEM260, TMEM43, TMEM70, TNNC1, TNNI3, TNNT2, TPM1, TRDN, TRPM4, TSFM, TTN, TTR, VCL, ZEB2, ZFPM2, ZIC3, ZNF469
Technical Specifications
● Genomic regions not covered by this test:
• NM_001374258.1 (BRAF): exon 10 and exon 20
• NM_001042492.3 (NF1): exon 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,28, 29, 32, 33, 34, 35 and 36
• NM_005027.4 (PIK3R2): exon 6
• NM_000384.3 (APOB): exon 1
• NM_015627.3 (LDLRAP1): exon 1
• NM_000237.3 (LPL): exon 5
• NM_014244.5 (ADAMTS2): exon 1
• NM_000722.4 (CACNA2D): exon 14
• NM_000071.3 (CBS): exon 15
• NM_000089.4 (COL1A2): exon 1
• NM_000093.5/NM_001278074.1 (COL5A1): exon 1
• NM_024757.5 (EHMT1): exon 1
• NM_153717.3 (EVC): exon 1
• NM_017617.5 (NOTCH1): exon 1
• NM_016203.4 (PRKAG2): exon 13
• NM_001037.5 (SCN1B): exon 1
• NM_004612.4 (TGFBR1): exon 1
• NM_006073.4 (TRDN): exon 5, 26, and 18
• NM_001267550.2 (TTN): exon 149, 164, 177, 186, 195
• NM_012082.4 (ZFPM2): exon 1
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Neonatal Panel analyzes 140 genes in symptomatic and pre-symptomatic infants.
Genes Tested
ABCD1, ABCD4, ACAD8, ACADM, ACADS, ACADSB, ACADVL, ACAT1, ADA, AGL, AGXT, AHCY, ALDOB, ALPL, AQP2, ARG1, ARSA, ASL, ASS1, ATP7B, AUH, BCKDHA, BCKDHB, BTD, CBS, CDH23, CFTR, CPT1A, CPT2, CTNS, CYP11B1, CYP11B2, CYP17A1, CYP27A1, DBT, DLD, ETFA, ETFB, ETFDH, FAH, G6PC, G6PD, GAA, GALC, GALE, GALK1, GALT, GAMT, GATM, GCDH, GCH1, GJB2, GJB6, GLA, GNMT, GRHPR, GYS2, HADH, HADHA, HADHB, HBB, HCFC1, HLCS, HMGCL, HOGA1, HPD, HSD17B10, HSD3B2, IDS, IDUA, IL2RG, IL7R, IVD, JAK3, KCNE1, KCNQ1, LDLR, LHX3, LIPA, LMBRD1, LPL, MAT1A, MCCC1, MCCC2, MCEE, MLYCD, MMAA, MMAB, MMACHC, MMADHC, MPI, MTR, MTRR, MTTP, MUT, MYO15A, NADK2, NAGS, NPC1, OTC, OTOF, PAH, PAX3, PAX8, PCCA, PCCB, PHGDH, PIK3CD, POR, PROP1, PYGL, SLC22A5, SLC25A13, SLC25A15, SLC25A20, SLC26A4, SLC37A4, SLC5A5, SLC7A7, SMPD1, SOX10, STAR, TAT, TECTA, TG, TH, THRA, TMIE, TMPRSS3, TPO, TPRN, TRIOBP, TRMU, TSHB, TSHR, UGT1A1, USH1C, USH1G, USH2A, WHRN
Technical Specifications
● Genomic regions not covered by this test:
• NM_000218.3 (KCNQ1): exon 1
• NM_001085411.3 (NADK2): exon 1
• NM_014564.5 (LHX3): exon 1
• NM_000156.6/NM_138924.3 (GAMT): exon 1
• NM_000402.4 (G6PD): exon 1
• NM_001352514.2 (HLCS): exon 1
• NM_012213.3 (MLYCD): exon 1
• NM_000203.5 (IDUA): exons 1 and 9
• NM_153006.3 (NAGS): exon 1
• NM_000022.4 (ADA): exon 1
• NM_147196.3 (TMIE): exon 1
• NM_000019.4 (ACAT1): exon 1
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Infertility Panel offers a female and a male infertility panel. The female infertility panel covers 54 genes, and the male infertility panel covers 39 genes. Structural and numerical abnormalities on sex chromosomes are included in both panels.
Genes Tested in Female Panel
AIRE, ANOS1, BMP15, CAPN10, CHD7, CYP11A1, CYP17A1, CYP19A1, DENND1A, DUSP6, EIF2B2, EIF2B3, FEZF1, FGF17, FGF8, FGFR1, FIGLA, FLRT3, FOXL2, FSHB, FSHR, GALT, GDF9, GNAS, GNRH1, GNRHR, HESX1, HS6ST1, IL17RD, INS, INSR, IRS1, IRS2, KISS1, KISS1R, LHB, LHCGR, NOBOX, NR5A1, NSMF, POF1B, POLG, PROK2, PROKR2, PSMC3IP, SEMA3A, SPRY4, STAG3, TAC3, TACR3, THADA, WDR11, WT1, ZP1
Genes Tested in Male Panel
ANOS1, AR, AURKC, CATSPER1, CFTR, CHD7, DAZL, DDX25, DUSP6, FEZF1, FGF17, FGF8, FGFR1, FLRT3, FSHB, FSHR, GNRH1, GNRHR, HESX1, HS6ST1, IL17RD, KISS1, KISS1R, LHB, LHCGR, NR5A1, NSMF, PRM1, PROK2, PROKR2, SEMA3A, SPRY4, SRD5A1, SRY, TAC3, TACR3, USP26, USP9Y, WDR11
Technical Specifications for Female Panel
● Genomic regions not covered by this test:
• NM_001282717.2 (STAG3): exon 7
• NM_001004311.3 (FIGLA): exons 1 and 4
• NM_004807.3 (HS6ST1): exon 2
• NM_001352964.2 (DENND1A): exons 1, 20 and 22
• NM_000208.4 (INSR): exon 1
• NM_033163.5 (FGF8): exon 1
• NM_000894.3 (LHB): exon 1
• NM_001130969.3 (NSMF): exon 1
• NM_001126128.2 (PROK2): exon 1
Technical Specifications for Male Panel
● Genomic regions not covered by this test:
• NM_004807.3 (HS6ST1): exon 2
• NM_001351.4 (DAZL): exons 7, 9, 10 and 11
• NM_033163.5 (FGF8): exon 1
• NM_000894.3 (LHB): exon 1
• NM_000894.3 (LHB): exon 1
• NM_000894.3 (LHB): exon 1
Technical Limitations for Female and Male Panels
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of extreme GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Sex chromosomal numerical and structural abnormalities (aneuploidies, copy number changes ≥10Mb, and mosaicism ≥15%) can be detected by Rodinia test. Although the test is highly accurate there is still a possibility for false positive and false negative results.
Description
Hereditary Cancer Panel analyzes 62 genes and covers 24 cancer predisposing syndromes associated with hereditary cancer.
Genes Tested
APC, ATM, BAP1, BARD1, BMPR1A, BRCA1, BRCA2, BRIP1, CDH1, CDK4, CDKN2A, CHEK2, DDB2, DICER1, EPCAM, ERCC1, ERCC2, ERCC3, ERCC4, ERCC5, FANCA, FANCB, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCL, FANCM, GREM1, HOXB13, MEN1, MLH1, MRE11, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, POLD1, POLE, POLH, PTEN, RAD50, RAD51C, RAD51D, RB1, RET, SDHAF2, SDHB, SDHC, SDHD, SLX4, SMAD4, SMARCA4, STK11, TP53, VHL, XPA, XPC
Technical Specifications
● APC(NM_000038.6): Selected region of promoter 1B is covered for sequence changes and selected region of promoter 1A is covered for both sequence changes and CNVs.
● CDKN2A: Analysis is performed for transcripts NM_000077.5 and NM_058195.4 that encode the p16INK4a and p14ARF proteins respectively.
● EPCAM(NM_002354.3): Analysis is limited to CNV analysis.
● GREM1(NM_013372.7): Analysis is limited to CNV analysis of the 40 kb 5’ upstream regulatory region.
● HOXB13: Analysis is limited to NM_006361.6: c.251G>A (p.G84E) variant.
● MLH1(NM_000249.4): Selected region of the promoter is covered for sequence changes.
● MSH2(NM_000251.3): Selected region of the promoter is covered for sequence changes.
● PTEN(NM_000314.8): Selected region of the promoter is covered for both sequence changes and CNVs.
● TP53(NM_000546.6): Selected region of the promoter is covered for CNVs.
● Genomic regions not covered by this test:
• BMPR1A (NM_004329.3): exon 13
• CHEK2 (NM_007194.4): exons 12-15
• DICER1 (NM_177438.3): exon 27
• FANCA (NM_000135.4): exon 15
• FANCD2 (NM_001018115.30): exons 12-17 and 19-28
• PMS2 (NM_000535.7): exons 11-15
• POLE (NM_006231.4): exon 1
• PTEN (NM_000314.8): exon 9
• RB1 (NM_000321.3): exon 15
• SDHC (NM_003001.5): exon 6
• SMARCA4 (NM_001387283.1): exons 27, 30
Technical Limitations
This test aims to detect all variants relevant to hereditary cancer predisposition in the genes (MANE and/or canonical transcripts) listed above, by targeting the exons, 20 bp of adjacent intronic sequence, selected untranslated regions (UTR) and promoter regions. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other deep intronic regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low capability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Carrier Screening Comprehensive Panel analyzes 228 genes associated with genetic diseases that have moderate to severe well-defined phenotype and high cumulative frequency in individuals with unknown carrier status.
Genes Tested
ABCD1, ACAD9, ACADM, ACOX1, ACSF3, ADAMTS2, AGA, AGL, AGPS, AGXT, AIRE, ALDH3A2, ALDOB, ALG6, ALMS1, ALPL, AMT, AQP2, ARSA, ASL, ASNS, ASPA, ASS1, ATM, ATP6V1B1, ATP7B, BBS1, BBS12, BCKDHB, BCS1L, BLM, BSND, BTD, CAPN3, CBS, CEP290, CERKL, CFTR, CHM, CHRNE, CIITA, CLN3, CLN5, CLN6, CLN8, CLRN1, CNGB3, COL4A3, COL4A5, COL7A1, CPT1A, CPT2, CRB1, CTNS, CTSK, CYBB, CYP11B2, CYP19A1, CYP27A1, DCLRE1C, DHCR7, DHDDS, DLD, DMD, DNAH5, DNAI1, DNAI2, DYSF, EDA, EIF2B5, EMD, ESCO2, ETFA, ETHE1, EYS, F11, F5, F9, FAH, FAM161A, FANCC, FANCG, G6PC, GAA, GALC, GALK1, GALT, GBA, GBE1, GCDH, GFM1, GJB2, GJB6, GLA, GLDC, GNE, GNPTAB, GNPTG, GNS, GRHPR, HADHA, HAX1, HBA1, HBA2, HBB, HEXA, HEXB, HJV, HGSNAT, HLCS, HMGCL, HOGA1, HPS1, HPS3, HSD17B4, HYAL1, HYLS1, IDS, ELP1, IL2RG, IVD, LAMC2, LCA5, LDLR, LHCGR, LHX3, LIFR, LIPA, LOXHD1, LPL, LRPPRC, MCCC1, MCCC2, MCOLN1, MEFV, MFSD8, MKS1, MLC1, MMAA, MMAB, MMACHC, MMADHC, MPI, MPV17, MTM1, MTRR, MTTP, MMUT, NAGLU, NAGS, NBN, NDUFAF5, NDUFS6, NPC1, NPC2, NPHS1, NPHS2, NR2E3, NTRK1, OAT, OPA3, OTC, PAH, PCDH15, PDHA1, PDHB, PEX1, PEX10, PEX2, PEX6, PEX7, PFKM, PHGDH, PKHD1, PMM2, PPT1, PROP1, PSAP, PTS, PUS1, PYGM, RAB23, RAG2, RAPSN, RARS2, RLBP1, RMRP, RPGR, RS1, SACS, SAMHD1, SEPSECS, SGCA, SGCB, SGCG, SLC12A6, SLC17A5, SLC25A13, SLC25A15, SLC26A2, SLC26A4, SLC35A3, SLC37A4, SLC4A11, SLC6A8, SLC7A7, SMARCAL1, SMPD1, STAR, SUMF1, TFR2, TGM1, TH, TMEM216, TPP1, TRMU, TSFM, TTPA, TYMP, UGT1A1, USH1C, USH2A, VPS13A, VPS45, VPS53, VRK1, VSX2, WNT10A
Technical Specifications
● DMD: Deletion/Duplication analysis at single exon resolution is not offered for NM_004006.3 (DMD): exon 2, exon 15, exon 22, exon 23, exon 26, exon 27, exon 28, exon 32, exon 33, exon 34, exon 36, exon 38, exon 39, exon 40, exon 41, exon 42, exon 46, exon 54, exon 56, exon 58, exon 67, exon 68, exon 69, exon 71, exon 72, exon 73, exon 77, exon 78, exon 79.
● HBA1/2: the assay is designed to detect deletions and duplications of HBA1 and HBA2. All positive Copy Number Variations (CNVs) are confirmed using an orthogonal method. The test does not distinguish among the different HBA types of deletions/duplications, and any variant identified will be called as HBA loss/gain.
● Genomic regions not covered by this test:
• NM_000383.4 (AIRE): exon 1
• NM_000414.4 (HSD17B4): exon 5
• NM_001352514.2 (HLCS): exon 1
• NM_013339.4 (ALG6): exon 13
• NM_014244.5 (ADAMTS2): exon 1
• NM_025114.4 (CEP290): exons 5 and 23
• NM_000233.4 (LHCGR): exon 1
• NM_001384140.1 (PCDH15): exons 33, 34 and 35
• NM_153818.2/NM_002617.4 (PEX10): exon 1
• NM_032520.5 (GNPTG): exon 1
• NM_000521.4 (HEXB): exon 4
• NM_000232.5 (SGCB): exon 1
• NM_020533.3 (MCOLN1): exon 1
• NM_152419.3 (HGSNAT): exon 1
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Carrier Screening Core Panel analyzes 19 genes associated with genetic diseases of high incidence and severity in individuals with unknown carrier status.
Genes Tested
ACADM, ASPA, BLM, CFTR, DHCR7, DMD, ELP1, FANCC, GALT, GBA, GJB2, GJB6, HBA1, HBA2, HBB, HEXA, MCOLN1, PAH, SMPD1
Technical Specifications
● DMD: Deletion/Duplication analysis at single exon resolution is not offered for NM_004006.3 (DMD): exon 1, exon 2, exon 23, exon 25, exon 26, exon 27, exon 28, exon 31, exon 33, exon 34, exon 35, exon 36, exon 38, exon 39, exon 40, exon 41, exon 42, exon 54, exon 58, exon 60, exon 67, exon 69, exon 71, exon 72, exon 73, exon 75, exon 77, exon 78.
● HBA1/2: the assay is designed to detect deletions and duplications of HBA1 and HBA2. All positive Copy Number Variations (CNVs) are confirmed using an orthogonal method. The test does not distinguish among the different HBA types of deletions/duplications, and any variant identified will be called as HBA loss/gain.
● Genomic regions not covered by this test:
• NM_020533.3 (MCOLN1): exon 1
• NM_004006.3 (DMD): exon 23
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Metabolic Panel analyzes 223 genes and covers major classes of inherited metabolic disorders.
Genes Tested
ABCC8, ABCD1, ABCD4, ACAD8, ACAD9, ACADM, ACADS, ACADSB, ACADVL, ACOX1, ACSF3, AGA, AGL, AGPS, ALDH6A1, ALDOA, ALDOB, ALG1, ALG11, ALG12, ALG13, ALG2, ALG3, ALG6, ALG8, ALG9, AMACR, AMT, ARG1, ARSA, ARSB, ASL, ASPA, ASS1, AUH, B4GALT1, BCKDHA, BCKDHB, BTD, CAD, CCDC115, CD320, CLN3, CLN5, CLN6, CLN8, CLPB, COG1, COG2, COG4, COG5, COG6, COG7, COG8, CPS1, CPT1A, CPT2, CTNS, CTSA, CTSK, DBT, DDOST, DHCR7, DHDDS, DLD, DNAJC12, DNAJC19, DOLK, DPAGT1, DPM1, DPM2, DPM3, ENO3, ETFA, ETFB, ETFDH, FBP1, FUCA1, FUT8, G6PC, GAA, GALC, GALNS, GAMT, GATM, GBA, GBE1, GCDH, GCH1, GCK, GCSH, GLA, GLB1, GLDC, GLUD1, GM2A, GMPPA, GNE, GNPTAB, GNPTG, GNS, GUSB, GYG1, GYS1, GYS2, HADH, HADHA, HADHB, HCFC1, HEXA, HEXB, HGSNAT, HMGCL, HMGCS2, HPD, HRAS, HSD17B10, HSD17B4, HYAL1, IDS, IDUA, INSR, KCNJ11, LAMP2, LDHA, LIAS, LIPA, LMBRD1, MAN1B1, MAN2B1, MANBA, MCEE, MCOLN1, MFSD8, MGAT2, MLYCD, MMAA, MMAB, MMACHC, MMADHC, MMUT, MOGS, MPDU1, MPI, MTR, MTRR, NAGA, NAGLU, NAGS, NEU1, NGLY1, NPC1, NPC2, NUS1, OPA3, OTC, PAH, PCBD1, PEX1, PEX10, PEX11B, PEX12, PEX13, PEX14, PEX16, PEX19, PEX2, PEX26, PEX3, PEX5, PEX6, PEX7, PFKM, PGAM2, PGM1, PHKA1, PHKA2, PHKB, PHKG2, PHYH, PMM2, PPM1K, PPT1, PRKAG2, PSAP, PTS, PYGL, PYGM, QDPR, RFT1, SCP2, SERAC1, SGSH, SLC16A1, SLC17A5, SLC22A5, SLC25A13, SLC25A15, SLC25A20, SLC2A2, SLC35A1, SLC35A2, SLC35C1, SLC37A4, SLC39A8, SLC6A8, SLC6A9, SMPD1, SRD5A3, SSR4, STT3A, STT3B, SUCLA2, SUCLG1, SUMF1, TAFAZZIN, TIMM50, TMEM165, TMEM199, TMEM70, TPP1, TUSC3, VPS33A
Technical Specifications
● Genomic regions not covered by this test:
• NM_032520.5 (GNPTG): exon 1
• NM_152419.3 (HGSNAT): exon 1
• NM_000203.5 (IDUA): exon 9
• NM_000208.4 (INSR): exon 1
• NM_000263.4 (NAGLU): exon 1
• NM_000281.4 (PCBD1): exon 1
• NM_002617.4 (PEX10): exon 1
• NM_000288.4 (PEX7): exon 1
• NM_016203.4 (PRKAG2): exon 5
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
FH, PH and RAS Kit analyzes 11, 11 and 30 genes and covers Familial Hypercholesterolemia (FH), Pulmonary Hypertension (PH) and RASopathy (RAS) related disorders, respectively.
Genes Tested for FH
ABCA1, ABCG5, ABCG8, APOA5, APOB, APOE, LDLR, LDLRAP1, LIPA, LPL, PCSK9
Genes Tested for PH
ACVRL1, BMPR1B, BMPR2, CAV1, EIF2AK4, ENG, KCNA5, KCNK3, SMAD4, SMAD9, TBX4
Genes Tested for RASopathy
AKT3, BRAF, CBL, CCND2, EPHB4, HRAS, KRAS, LZTR1, MAP2K1, MAP2K2, MRAS, NF1, NF2, NRAS, PIK3CA, PIK3R2, PPP1CB, PTPN11, RAF1, RASA1, RASA2, RIT1, RRAS, SASH1, SHOC2, SMARCB1, SOS1, SOS2, SPRED1, STAMBP
Technical Specifications
● Genomic regions not covered by FH:
• NM_000384.3 (APOB): exon 1
• NM_015627.3 (LDLRAP1): exon 1
• NM_000237.3 (LPL): exon 5
● Genomic regions not covered by PH:
• Not Applicable
● Genomic regions not covered by RAS:
• NM_001374258.1 (BRAF): exon 10 and exon 20
• NM_001042492.3 (NF1): exon 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,28, 29, 32, 33, 34, 35 and 36
• NM_005027.4 (PIK3R2): exon 6
Technical Limitations for FH, PH and RAS
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
PGT Kit detects whole chromosome aneuploidies, structural rearrangements and segmental aneuploidies down to 10Mb, selected male polyploidies, and mosaicism higher than 50%.
Medicover Genetics offers on-going support to all Technology Transfer partners through dedicated customer service channels. Members of our team are always available to offer their assistance for troubleshooting, quality and performance monitoring, marketing and commercialization insights and technical support. Our provided software is updated automatically to ensure seamless operations.
Medicover Genetics Technology Transfer platform and software provide easy to interpret results through visualization in graphical and tubular format, instead of raw data. We also offer template reports and guidance upon request to make it easier for each laboratory to create their own reports.
POLARIS is a genetic data management system that enables laboratory personnel to manage sample data generated during the VERACITY and VERACITY High Content (HC) workflow. POLARIS interfaces with the Medicover Genetics analysis engine VEGA, to provide information for the analysis runs and retrieve the analysis data for visualization in graphical and tabular format. POLARIS can also be integrated to local Laboratory Management Systems.
SIRIUS is a genetic data management web application that enables users to manage information generated by NGS analysis. Additionally, SIRIUS facilitates the creation, calculation, and modification of sample batches corresponding to sequencing runs and interfaces with the Medicover Genetics analysis engine VEGA, to provide information on the analysis data in visual graphs and tabular formats.
Description
Metabolic Kit analyzes 223 genes and covers major classes of inherited metabolic disorders.
Genes Tested
ABCC8, ABCD1, ABCD4, ACAD8, ACAD9, ACADM, ACADS, ACADSB, ACADVL, ACOX1, ACSF3, AGA, AGL, AGPS, ALDH6A1, ALDOA, ALDOB, ALG1, ALG11, ALG12, ALG13, ALG2, ALG3, ALG6, ALG8, ALG9, AMACR, AMT, ARG1, ARSA, ARSB, ASL, ASPA, ASS1, AUH, B4GALT1, BCKDHA, BCKDHB, BTD, CAD, CCDC115, CD320, CLN3, CLN5, CLN6, CLN8, CLPB, COG1, COG2, COG4, COG5, COG6, COG7, COG8, CPS1, CPT1A, CPT2, CTNS, CTSA, CTSK, DBT, DDOST, DHCR7, DHDDS, DLD, DNAJC12, DNAJC19, DOLK, DPAGT1, DPM1, DPM2, DPM3, ENO3, ETFA, ETFB, ETFDH, FBP1, FUCA1, FUT8, G6PC, GAA, GALC, GALNS, GAMT, GATM, GBA, GBE1, GCDH, GCH1, GCK, GCSH, GLA, GLB1, GLDC, GLUD1, GM2A, GMPPA, GNE, GNPTAB, GNPTG, GNS, GUSB, GYG1, GYS1, GYS2, HADH, HADHA, HADHB, HCFC1, HEXA, HEXB, HGSNAT, HMGCL, HMGCS2, HPD, HRAS, HSD17B10, HSD17B4, HYAL1, IDS, IDUA, INSR, KCNJ11, LAMP2, LDHA, LIAS, LIPA, LMBRD1, MAN1B1, MAN2B1, MANBA, MCEE, MCOLN1, MFSD8, MGAT2, MLYCD, MMAA, MMAB, MMACHC, MMADHC, MMUT, MOGS, MPDU1, MPI, MTR, MTRR, NAGA, NAGLU, NAGS, NEU1, NGLY1, NPC1, NPC2, NUS1, OPA3, OTC, PAH, PCBD1, PEX1, PEX10, PEX11B, PEX12, PEX13, PEX14, PEX16, PEX19, PEX2, PEX26, PEX3, PEX5, PEX6, PEX7, PFKM, PGAM2, PGM1, PHKA1, PHKA2, PHKB, PHKG2, PHYH, PMM2, PPM1K, PPT1, PRKAG2, PSAP, PTS, PYGL, PYGM, QDPR, RFT1, SCP2, SERAC1, SGSH, SLC16A1, SLC17A5, SLC22A5, SLC25A13, SLC25A15, SLC25A20, SLC2A2, SLC35A1, SLC35A2, SLC35C1, SLC37A4, SLC39A8, SLC6A8, SLC6A9, SMPD1, SRD5A3, SSR4, STT3A, STT3B, SUCLA2, SUCLG1, SUMF1, TAFAZZIN, TIMM50, TMEM165, TMEM199, TMEM70, TPP1, TUSC3, VPS33A
Technical Specifications
● Genomic regions not covered by this test:
• NM_032520.5 (GNPTG): exon 1
• NM_152419.3 (HGSNAT): exon 1
• NM_000203.5 (IDUA): exon 9
• NM_000208.4 (INSR): exon 1
• NM_000263.4 (NAGLU): exon 1
• NM_000281.4 (PCBD1): exon 1
• NM_002617.4 (PEX10): exon 1
• NM_000288.4 (PEX7): exon 1
• NM_016203.4 (PRKAG2): exon 5
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Congenital Heart Defects Kit analyzes 80 genes and covers inherited congenital heart defects.
Genes Tested
ACTA2, ACTC1, ACVR2B, AFF4, BMPR2, CAD, CFAP53, CDK13, CELSR1, CELSR2, CELSR3, CHD4, CHD7, CITED2, CREBBP, CRELD1, DNAH11, DNAH5, DNAH6, DNAI1, DTNA, EHMT1, ELN, EVC, EVC2, FBN1, FLNA, FOXC1, FOXH1, GANAB, GATA4, GATA5, GATA6, GDF1, GPC3, HAND1, HAND2, HRAS, JAG1, KDM5B, KMT2D, MED13L, MMP21, MYH11, MYH6, MYH7, NIPBL, NKX2-5, NKX2-6, NME7, NODAL, NOTCH1, NOTCH2, NR2F2, PITX2, PKD1L1, PLD1, POGZ, PRDM6, PRKD1, RABGAP1L, RBFOX2, RBM10, SALL4, SEMA3D, SEMA3E, SMAD6, TAB2, TBX1, TBX20, TBX5, TFAP2B, TGFBR1, TGFBR2, TLL1, TMEM260, TPM1, ZEB2, ZFPM2, ZIC3
Technical Specifications
● Genomic regions not covered by this test:
• NM_005159.5 (ACTC1): exon 2
• NM_001106.4 (ACVR2B): exon 1
• NM_024757.5 (EHMT1): exon 1
• NM_153717.3 (EVC): exon 1
• NM_001110556.2 (FLNA): exon 15
• NM_001308093.3 (GATA4): exon 1
• NM_001492.6 (GDF1): exon 8
• NM_000214.3 (JAG1): exon 1
• NM_017617.5 (NOTCH1): exon 1
• NM_001136239.4 (PRDM6): exon 2
• NM_001349999.2 (RBFOX2): exon 1
• NM_005585.5 (SMAD6): exon 1
• NM_080647.1 (TBX1): exon 8, and 9
• NM_004612.4 (TGFBR1): exon 1
• NM_012082.4 (ZFPM2): exon 1
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Aortopathy Kit analyzes 48 genes and covers aortopathy-related inherited cardiovascular disorders.
Genes Tested
ABCC6, ACTA2, ACVR1, ADAMTS2, ALDH18A1, ATP6V0A2, ATP6V1A, ATP6V1E1, B3GALT6, B4GALT7, BGN, C1R, C1S, CBS, CHST14, COL1A1, COL1A2, COL3A1, COL4A1, COL5A1, COL5A2, DSE, EFEMP2, ELN, FBLN5, FBN1, FBN2, FKBP14, FOXE3, GORAB, LOX, LTBP4, MFAP5, MYH11, MYLK, PLOD1, PRDM5, PYCR1, SKI, SLC2A10, SLC39A13, SMAD3, SMAD4, TGFB2, TGFB3, TGFBR1, TGFBR2, ZNF469
Technical Specifications
● Genomic regions not covered by this test:
• NM_014244.5 (ADAMTS2): exon 1
• NM_007255.3 (B4GALT7): exon 1
• NM_000071.3 (CBS): exon 15
• NM_000089.4 (COL1A2): exon 3
• NM_001278074.1 (COL5A1): exon 1
• NM_001042545.2 (LTBP4): exon 4
• NM_004612.4 (TGFBR1): exon 1
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Arrhythmia Kit analyzes 42 genes and covers arrhythmia-related inherited cardiovascular disorders.
Genes Tested
ABCC9, AKAP9, ANK2, CACNA1C, CACNA2D1, CACNB2, CALM1, CALM2, CALM3, CASQ2, CAV3, GJA5, GNAI2, GPD1L, HCN4, KCNA5, KCNAB2, KCND3, KCNE1, KCNE2, KCNE3, KCNH2, KCNJ2, KCNJ5, KCNJ8, KCNQ1, LMNA, MYL4, NPPA, RYR2, SCN10A, SCN1B, SCN2B, SCN3B, SCN4B, SCN5A, SNTA1, TBX5, TECRL, TNNI3, TRDN, TRPM4
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Cardiomyopathy Kit analyzes 98 genes and covers cardiomyopathy-related inherited cardiovascular disorders.
Genes Tested
AARS2, ABCC9, ACAD9, ACADVL, ACTA1, ACTC1, ACTN2, AGK, AKAP9, ALMS1, ALPK3, ANKRD1, BAG3, CASQ2, CAV3, COA5, COA6, COX15, CPT2, CRYAB, CSRP3, CTNNA3, DES, DMD, DNAJC19, DOLK, DSC2, DSG2, DSP, DTNA, ELAC2, EMD, FHL1, FKTN, FLNC, GAA, GATAD1, GBE1, GLA, GTPBP3, HADHA, HADHB, HCN4, HRAS, ILK, JPH2, JUP, KARS1, LAMA4, LAMP2, LDB3, LMNA, MIB1, MRPL3, MRPL44, MTO1, MYBPC3, MYH6, MYH7, MYL2, MYL3, MYLK2, MYOZ2, MYPN, NDUFB11, NDUFV2, NEBL, NEXN, PDLIM3, PKP2, PLN, PPA2, PRDM16, PRKAG2, RAF1, RBM20, RYR2, SCN5A, SCO2, SGCD, SLC22A5, SLC25A20, SLC25A3, SLC25A4, TAFAZZIN, TCAP, TGFB3, TK2, TMEM43, TMEM70, TNNC1, TNNI3, TNNT2, TPM1, TSFM, TTN, TTR, VCL
Technical Specifications
● Genomic regions not covered by this test:
• NM_000018.4 (ACADVL): exon 2
• NM_001943.5 (DSG2): exon 1
• NM_001458.5 (FLNC): exon 20
• NM_021167.5 (GATAD1): exon 1
• NM_020433.5 (JPH2): exon 4
• NM_002230.4 (JUP): exon 11
• NM_016203.4 (PRKAG2): exon 13
• NM_014000.3 (VCL): exon 11
• NM_001267550.2 (TTN): exon 149, 164, 175, 176, 177, 184, 186, 193, 194, 195, 215
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Cardiac Comprehensive Kit analyzes 292 genes and covers major inherited cardiovascular disorders.
Genes Tested
AARS2, ABCA1, ABCC6, ABCC9, ABCG5, ABCG8, ACAD9, ACADVL, ACTA1, ACTA2, ACTC1, ACTN2, ACVR1, ACVR2B, ACVRL1, ADAMTS2, AFF4, AGK, AKAP9, AKT3, ALDH18A1, ALMS1, ALPK3, ANK2, ANKRD1, APOA5, APOB, APOE, ATP6V0A2, ATP6V1A, ATP6V1E1, B3GALT6, B4GALT7, BAG3, BGN, BMPR1B, BMPR2, BRAF, C1R, C1S, CACNA1C, CACNA2D1, CACNB2, CAD, CALM1, CALM2, CALM3, CASQ2, CAV1, CAV3, CBL, CBS, CFAP53, CCND2, CDK13, CELSR1, CELSR2, CELSR3, CHD4, CHD7, CHST14, CITED2, COA5, COA6, COL1A1, COL1A2, COL3A1, COL4A1, COL5A1, COL5A2, COX15, CPT2, CREBBP, CRELD1, CRYAB, CSRP3, CTNNA3, DES, DMD, DNAH11, DNAH5, DNAH6, DNAI1, DNAJC19, DOLK, DSC2, DSE, DSG2, DSP, DTNA, EFEMP2, EHMT1, EIF2AK4, ELAC2, ELN, EMD, ENG, EPHB4, EVC, EVC2, FBLN5, FBN1, FBN2, FHL1, FKBP14, FKTN, FLNA, FLNC, FOXC1, FOXE3, FOXH1, GAA, GANAB, GATA4, GATA5, GATA6, GATAD1, GBE1, GDF1, GJA5, GLA, GNAI2, GORAB, GPC3, GPD1L, GTPBP3, HADHA, HADHB, HAND1, HAND2, HCN4, HRAS, ILK, JAG1, JPH2, JUP, KARS1, KCNA5, KCNAB2, KCND3, KCNE1, KCNE2, KCNE3, KCNH2, KCNJ2, KCNJ5, KCNJ8, KCNK3, KCNQ1, KDM5B, KMT2D, KRAS, LAMA4, LAMP2, LDB3, LDLR, LDLRAP1, LIPA, LMNA, LOX, LPL, LTBP4, LZTR1, MAP2K1, MAP2K2, MED13L, MFAP5, MIB1, MMP21, MRAS, MRPL3, MRPL44, MTO1, MYBPC3, MYH11, MYH6, MYH7, MYL2, MYL3, MYL4, MYLK, MYLK2, MYOZ2, MYPN, NDUFB11, NDUFV2, NEBL, NEXN, NF1, NF2, NIPBL, NKX2-5, NKX2-6, NME7, NODAL, NOTCH1, NOTCH2, NPPA, NR2F2, NRAS, PCSK9, PDLIM3, PIK3CA, PIK3R2, PITX2, PKD1L1, PKP2, PLD1, PLN, PLOD1, POGZ, PPA2, PPP1CB, PRDM16, PRDM5, PRDM6, PRKAG2, PRKD1, PTPN11, PYCR1, RABGAP1L, RAF1, RASA1, RASA2, RBFOX2, RBM10, RBM20, RIT1, RRAS, RYR2, SALL4, SASH1, SCN10A, SCN1B, SCN2B, SCN3B, SCN4B, SCN5A, SCO2, SEMA3D, SEMA3E, SGCD, SHOC2, SKI, SLC22A5, SLC25A20, SLC25A3, SLC25A4, SLC2A10, SLC39A13, SMAD3, SMAD4, SMAD6, SMAD9, SMARCB1, SNTA1, SOS1, SOS2, SPRED1, STAMBP, TAB2, TAFAZZIN, TBX1, TBX20, TBX4, TBX5, TCAP, TECRL, TFAP2B, TGFB2, TGFB3, TGFBR1, TGFBR2, TK2, TLL1, TMEM260, TMEM43, TMEM70, TNNC1, TNNI3, TNNT2, TPM1, TRDN, TRPM4, TSFM, TTN, TTR, VCL, ZEB2, ZFPM2, ZIC3, ZNF469
Technical Specifications
● Genomic regions not covered by this test:
• NM_001374258.1 (BRAF): exon 10 and exon 20
• NM_001042492.3 (NF1): exon 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,28, 29, 32, 33, 34, 35 and 36
• NM_005027.4 (PIK3R2): exon 6
• NM_000384.3 (APOB): exon 1
• NM_015627.3 (LDLRAP1): exon 1
• NM_000237.3 (LPL): exon 5
• NM_014244.5 (ADAMTS2): exon 1
• NM_000722.4 (CACNA2D): exon 14
• NM_000071.3 (CBS): exon 15
• NM_000089.4 (COL1A2): exon 1
• NM_000093.5/NM_001278074.1 (COL5A1): exon 1
• NM_024757.5 (EHMT1): exon 1
• NM_153717.3 (EVC): exon 1
• NM_017617.5 (NOTCH1): exon 1
• NM_016203.4 (PRKAG2): exon 13
• NM_001037.5 (SCN1B): exon 1
• NM_004612.4 (TGFBR1): exon 1
• NM_006073.4 (TRDN): exon 5, 26, and 18
• NM_001267550.2 (TTN): exon 149, 164, 177, 186, 195
• NM_012082.4 (ZFPM2): exon 1
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Neonatal kit analyzes 140 genes in symptomatic and pre-symptomatic infants.
Genes Tested
ABCD1, ABCD4, ACAD8, ACADM, ACADS, ACADSB, ACADVL, ACAT1, ADA, AGL, AGXT, AHCY, ALDOB, ALPL, AQP2, ARG1, ARSA, ASL, ASS1, ATP7B, AUH, BCKDHA, BCKDHB, BTD, CBS, CDH23, CFTR, CPT1A, CPT2, CTNS, CYP11B1, CYP11B2, CYP17A1, CYP27A1, DBT, DLD, ETFA, ETFB, ETFDH, FAH, G6PC, G6PD, GAA, GALC, GALE, GALK1, GALT, GAMT, GATM, GCDH, GCH1, GJB2, GJB6, GLA, GNMT, GRHPR, GYS2, HADH, HADHA, HADHB, HBB, HCFC1, HLCS, HMGCL, HOGA1, HPD, HSD17B10, HSD3B2, IDS, IDUA, IL2RG, IL7R, IVD, JAK3, KCNE1, KCNQ1, LDLR, LHX3, LIPA, LMBRD1, LPL, MAT1A, MCCC1, MCCC2, MCEE, MLYCD, MMAA, MMAB, MMACHC, MMADHC, MPI, MTR, MTRR, MTTP, MUT, MYO15A, NADK2, NAGS, NPC1, OTC, OTOF, PAH, PAX3, PAX8, PCCA, PCCB, PHGDH, PIK3CD, POR, PROP1, PYGL, SLC22A5, SLC25A13, SLC25A15, SLC25A20, SLC26A4, SLC37A4, SLC5A5, SLC7A7, SMPD1, SOX10, STAR, TAT, TECTA, TG, TH, THRA, TMIE, TMPRSS3, TPO, TPRN, TRIOBP, TRMU, TSHB, TSHR, UGT1A1, USH1C, USH1G, USH2A, WHRN
Technical Specifications
● Genomic regions not covered by this test:
• NM_000218.3 (KCNQ1): exon 1
• NM_001085411.3 (NADK2): exon 1
• NM_014564.5 (LHX3): exon 1
• NM_000156.6/NM_138924.3 (GAMT): exon 1
• NM_000402.4 (G6PD): exon 1
• NM_001352514.2 (HLCS): exon 1
• NM_012213.3 (MLYCD): exon 1
• NM_000203.5 (IDUA): exons 1 and 9
• NM_153006.3 (NAGS): exon 1
• NM_000022.4 (ADA): exon 1
• NM_147196.3 (TMIE): exon 1
• NM_000019.4 (ACAT1): exon 1
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Infertility Kit offers a female and a male infertility panel. The female infertility panel covers 54 genes, and the male infertility panel covers 39 genes. Structural and numerical abnormalities on sex chromosomes are included in both panels.
Genes Tested in Female Panel
AIRE, ANOS1, BMP15, CAPN10, CHD7, CYP11A1, CYP17A1, CYP19A1, DENND1A, DUSP6, EIF2B2, EIF2B3, FEZF1, FGF17, FGF8, FGFR1, FIGLA, FLRT3, FOXL2, FSHB, FSHR, GALT, GDF9, GNAS, GNRH1, GNRHR, HESX1, HS6ST1, IL17RD, INS, INSR, IRS1, IRS2, KISS1, KISS1R, LHB, LHCGR, NOBOX, NR5A1, NSMF, POF1B, POLG, PROK2, PROKR2, PSMC3IP, SEMA3A, SPRY4, STAG3, TAC3, TACR3, THADA, WDR11, WT1, ZP1
Genes Tested in Male Panel
ANOS1, AR, AURKC, CATSPER1, CFTR, CHD7, DAZL, DDX25, DUSP6, FEZF1, FGF17, FGF8, FGFR1, FLRT3, FSHB, FSHR, GNRH1, GNRHR, HESX1, HS6ST1, IL17RD, KISS1, KISS1R, LHB, LHCGR, NR5A1, NSMF, PRM1, PROK2, PROKR2, SEMA3A, SPRY4, SRD5A1, SRY, TAC3, TACR3, USP26, USP9Y, WDR11
Technical Specifications for Female Panel
● Genomic regions not covered by this test:
• NM_001282717.2 (STAG3): exon 7
• NM_001004311.3 (FIGLA): exons 1 and 4
• NM_004807.3 (HS6ST1): exon 2
• NM_001352964.2 (DENND1A): exons 1, 20 and 22
• NM_000208.4 (INSR): exon 1
• NM_033163.5 (FGF8): exon 1
• NM_000894.3 (LHB): exon 1
• NM_001130969.3 (NSMF): exon 1
• NM_001126128.2 (PROK2): exon 1
Technical Specifications for Male Panel
● Genomic regions not covered by this test:
• NM_004807.3 (HS6ST1): exon 2
• NM_001351.4 (DAZL): exons 7, 9, 10 and 11
• NM_033163.5 (FGF8): exon 1
• NM_000894.3 (LHB): exon 1
• NM_000894.3 (LHB): exon 1
• NM_000894.3 (LHB): exon 1
Technical Limitations for Female and Male Panels
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of extreme GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Sex chromosomal numerical and structural abnormalities (aneuploidies, copy number changes ≥10Mb, and mosaicism ≥15%) can be detected by Rodinia test. Although the test is highly accurate there is still a possibility for false positive and false negative results.
Description
Hereditary Cancer Kit analyzes 62 genes and covers 24 cancer predisposing syndromes associated with hereditary cancer.
Genes Tested
APC, ATM, BAP1, BARD1, BMPR1A, BRCA1, BRCA2, BRIP1, CDH1, CDK4, CDKN2A, CHEK2, DDB2, DICER1, EPCAM, ERCC1, ERCC2, ERCC3, ERCC4, ERCC5, FANCA, FANCB, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCL, FANCM, GREM1, HOXB13, MEN1, MLH1, MRE11, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, POLD1, POLE, POLH, PTEN, RAD50, RAD51C, RAD51D, RB1, RET, SDHAF2, SDHB, SDHC, SDHD, SLX4, SMAD4, SMARCA4, STK11, TP53, VHL, XPA, XPC
Technical Specifications
● APC(NM_000038.6): Selected region of promoter 1B is covered for sequence changes and selected region of promoter 1A is covered for both sequence changes and CNVs.
● CDKN2A: Analysis is performed for transcripts NM_000077.5 and NM_058195.4 that encode the p16INK4a and p14ARF proteins respectively.
● EPCAM(NM_002354.3): Analysis is limited to CNV analysis.
● GREM1(NM_013372.7): Analysis is limited to CNV analysis of the 40 kb 5’ upstream regulatory region.
● HOXB13: Analysis is limited to NM_006361.6: c.251G>A (p.G84E) variant.
● MLH1(NM_000249.4): Selected region of the promoter is covered for sequence changes.
● MSH2(NM_000251.3): Selected region of the promoter is covered for sequence changes.
● PTEN(NM_000314.8): Selected region of the promoter is covered for both sequence changes and CNVs.
● TP53(NM_000546.6): Selected region of the promoter is covered for CNVs.
● Genomic regions not covered by this test:
• BMPR1A (NM_004329.3): exon 13
• CHEK2 (NM_007194.4): exons 12-15
• DICER1 (NM_177438.3): exon 27
• FANCA (NM_000135.4): exon 15
• FANCD2 (NM_001018115.30): exons 12-17 and 19-28
• PMS2 (NM_000535.7): exons 11-15
• POLE (NM_006231.4): exon 1
• PTEN (NM_000314.8): exon 9
• RB1 (NM_000321.3): exon 15
• SDHC (NM_003001.5): exon 6
• SMARCA4 (NM_001387283.1): exons 27, 30
Technical Limitations
This test aims to detect all variants relevant to hereditary cancer predisposition in the genes (MANE and/or canonical transcripts) listed above, by targeting the exons, 20 bp of adjacent intronic sequence, selected untranslated regions (UTR) and promoter regions. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other deep intronic regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low capability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Carrier Screening Comprehensive Kit analyzes 228 genes associated with genetic diseases that have moderate to severe well-defined phenotype and high cumulative frequency in individuals with unknown carrier status.
Genes Tested
ABCD1, ACAD9, ACADM, ACOX1, ACSF3, ADAMTS2, AGA, AGL, AGPS, AGXT, AIRE, ALDH3A2, ALDOB, ALG6, ALMS1, ALPL, AMT, AQP2, ARSA, ASL, ASNS, ASPA, ASS1, ATM, ATP6V1B1, ATP7B, BBS1, BBS12, BCKDHB, BCS1L, BLM, BSND, BTD, CAPN3, CBS, CEP290, CERKL, CFTR, CHM, CHRNE, CIITA, CLN3, CLN5, CLN6, CLN8, CLRN1, CNGB3, COL4A3, COL4A5, COL7A1, CPT1A, CPT2, CRB1, CTNS, CTSK, CYBB, CYP11B2, CYP19A1, CYP27A1, DCLRE1C, DHCR7, DHDDS, DLD, DMD, DNAH5, DNAI1, DNAI2, DYSF, EDA, EIF2B5, EMD, ESCO2, ETFA, ETHE1, EYS, F11, F5, F9, FAH, FAM161A, FANCC, FANCG, G6PC, GAA, GALC, GALK1, GALT, GBA, GBE1, GCDH, GFM1, GJB2, GJB6, GLA, GLDC, GNE, GNPTAB, GNPTG, GNS, GRHPR, HADHA, HAX1, HBA1, HBA2, HBB, HEXA, HEXB, HJV, HGSNAT, HLCS, HMGCL, HOGA1, HPS1, HPS3, HSD17B4, HYAL1, HYLS1, IDS, ELP1, IL2RG, IVD, LAMC2, LCA5, LDLR, LHCGR, LHX3, LIFR, LIPA, LOXHD1, LPL, LRPPRC, MCCC1, MCCC2, MCOLN1, MEFV, MFSD8, MKS1, MLC1, MMAA, MMAB, MMACHC, MMADHC, MPI, MPV17, MTM1, MTRR, MTTP, MMUT, NAGLU, NAGS, NBN, NDUFAF5, NDUFS6, NPC1, NPC2, NPHS1, NPHS2, NR2E3, NTRK1, OAT, OPA3, OTC, PAH, PCDH15, PDHA1, PDHB, PEX1, PEX10, PEX2, PEX6, PEX7, PFKM, PHGDH, PKHD1, PMM2, PPT1, PROP1, PSAP, PTS, PUS1, PYGM, RAB23, RAG2, RAPSN, RARS2, RLBP1, RMRP, RPGR, RS1, SACS, SAMHD1, SEPSECS, SGCA, SGCB, SGCG, SLC12A6, SLC17A5, SLC25A13, SLC25A15, SLC26A2, SLC26A4, SLC35A3, SLC37A4, SLC4A11, SLC6A8, SLC7A7, SMARCAL1, SMPD1, STAR, SUMF1, TFR2, TGM1, TH, TMEM216, TPP1, TRMU, TSFM, TTPA, TYMP, UGT1A1, USH1C, USH2A, VPS13A, VPS45, VPS53, VRK1, VSX2, WNT10A
Technical Specifications
● DMD: Deletion/Duplication analysis at single exon resolution is not offered for NM_004006.3 (DMD): exon 2, exon 15, exon 22, exon 23, exon 26, exon 27, exon 28, exon 32, exon 33, exon 34, exon 36, exon 38, exon 39, exon 40, exon 41, exon 42, exon 46, exon 54, exon 56, exon 58, exon 67, exon 68, exon 69, exon 71, exon 72, exon 73, exon 77, exon 78, exon 79.
● HBA1/2: the assay is designed to detect deletions and duplications of HBA1 and HBA2. All positive Copy Number Variations (CNVs) are confirmed using an orthogonal method. The test does not distinguish among the different HBA types of deletions/duplications, and any variant identified will be called as HBA loss/gain.
● Genomic regions not covered by this test:
• NM_000383.4 (AIRE): exon 1
• NM_000414.4 (HSD17B4): exon 5
• NM_001352514.2 (HLCS): exon 1
• NM_013339.4 (ALG6): exon 13
• NM_014244.5 (ADAMTS2): exon 1
• NM_025114.4 (CEP290): exons 5 and 23
• NM_000233.4 (LHCGR): exon 1
• NM_001384140.1 (PCDH15): exons 33, 34 and 35
• NM_153818.2/NM_002617.4 (PEX10): exon 1
• NM_032520.5 (GNPTG): exon 1
• NM_000521.4 (HEXB): exon 4
• NM_000232.5 (SGCB): exon 1
• NM_020533.3 (MCOLN1): exon 1
• NM_152419.3 (HGSNAT): exon 1
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
Description
Carrier Screening Core Kit analyzes 19 genes associated with genetic diseases of high incidence and severity in individuals with unknown carrier status.
Genes Tested
ACADM, ASPA, BLM, CFTR, DHCR7, DMD, ELP1, FANCC, GALT, GBA, GJB2, GJB6, HBA1, HBA2, HBB, HEXA, MCOLN1, PAH, SMPD1
Technical Specifications
● DMD: Deletion/Duplication analysis at single exon resolution is not offered for NM_004006.3 (DMD): exon 1, exon 2, exon 23, exon 25, exon 26, exon 27, exon 28, exon 31, exon 33, exon 34, exon 35, exon 36, exon 38, exon 39, exon 40, exon 41, exon 42, exon 54, exon 58, exon 60, exon 67, exon 69, exon 71, exon 72, exon 73, exon 75, exon 77, exon 78.
● HBA1/2: the assay is designed to detect deletions and duplications of HBA1 and HBA2. All positive Copy Number Variations (CNVs) are confirmed using an orthogonal method. The test does not distinguish among the different HBA types of deletions/duplications, and any variant identified will be called as HBA loss/gain.
● Genomic regions not covered by this test:
• NM_020533.3 (MCOLN1): exon 1
• NM_004006.3 (DMD): exon 23
Technical Limitations
The test aims to detect all variants relevant to the genes listed above by targeting all coding exons, of MANE or/and Canonical transcripts, and 12 bp of adjacent intronic sequence. Variants that fall outside of the targeted regions are not intended to be detected by this assay. Unless otherwise noted, sequence changes (SNVs and INDELS) in the promoter and other non-coding regions are not covered by this assay. Certain sequence changes (SNVs and INDELS) in non-coding regions of selected genes that are of clinical significance are also included in the analysis. In cases where two variants are identified in a gene, the test does not distinguish whether these are on one chromosome (in cis) or on different chromosomes (in trans). Certain types of genetic abnormalities such as inversions, rearrangements, polyploidy and epigenetic effects are not covered by this test. Certain sequence changes (SNVs and INDELS) in targeted regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of high/low GC-content may not be detected. Copy Number Variations (CNVs) are calculated using high quality, de-duplicated and uniquely aligned sequencing reads. CNVs are detected for a subset of the targeted regions using a depth of sequencing coverage approach by applying GC-content normalization. Genomic regions are called as variants if their normalized depth of coverage deviates significantly from the expected normalized coverage which is estimated from a set of reference clinical samples. The test can detect CNVs down to a few exons level resolution. The test cannot detect CNVs at genomic regions with either low mappability or containing repeats, pseudogenes and high/low GC-content. Detection of CNVs using NGS has lower sensitivity/specificity than orthogonal quantification methods, therefore the absence of reported CNVs does not guarantee the absence of CNVs. The lack of disease-causing variants in the targeted genes diminishes but does not exclude the possibility of a disease associated syndrome. Although the test is highly accurate there is still a possibility for false positive or false negative results.
We work closely with laboratories to meet their unique needs and goals. We understand that every laboratory is different, and we tailor our approach to accommodate the specific requirements of each partner. In collaboration with our partners, we design a seamless integrated process, from initial setup to ongoing support and maintenance. Our experienced team provides technical, commercial, and marketing assistance to ensure a successful partnership and optimal performance. We aim to build long-lasting partnerships that are mutually beneficial.

Medicover Genetics has created a straightforward forecast, order, and delivery plan to ensure on time delivery of reagents and uninterrupted operations of the laboratory. Logistics plan includes door-to-door delivery using temperature control packaging for assurance of product integrity.