Scientific Background
Ehlers-Danlos syndrome (EDS) refers to a clinically and genetically heterogeneous group of connective tissue disorders characterized by joint hypermobility, hyperextensible skin, and tissue fragility.
According to the simplified Villefranche classification of 1998, EDS was divided into six main subtypes based on clinical, biochemical and genetic data that are due to different molecular defects of collagen metabolism. Owing to the increase in genetically determined new EDS subtypes, the revised international classification of 2017 proposes 13 subtypes based on clinical criteria, which are inherited in an autosomal dominant or recessive pattern and, with the exception of the hypermobile EDS subtype, can be confirmed by genetic data. As a result of genetic causes and pathogenetic mechanisms, the individual EDS subtypes can be divided into different groups based on defects in collagen biosynthesis and processing, collagen folding and processing, the structure and function of the interface between muscle and the extracellular matrix, defects in glycosaminoglycan biosynthesis, the complement pathway, and intracellular processes.
Collagen biosynthesis and processing and the assembly of collagen fibrils is impaired at different levels in the EDS subtypes:
- (a) Synthesis and stability: haploinsufficiency of mutant COL5A1 mRNA leading to decreased synthesis of alpha-1-procollagen(V) is the cause in approximately 60% of classical EDS (cEDS).
- (b) Hydroxylation of lysine and proline in procollagen chains: lack of hydroxylation due to lysyl hydroxylase deficiency is the cause of kyphoscoliotic EDS (kEDS).
- (c) Processing and secretion: pathogenic variants in COL3A1 affecting the triple helix domain of procollagen alpha chains prevent normal processing in the rough endoplasmic reticulum and subsequent secretion of homotrimers. They are the cause of vascular EDS (vEDS).
- (d) Cleavage of N-terminal propeptides in the extracellular matrix: dominant pathogenic variants in COL1A1 and COL1A2 that prevent the recognition sequence cleaving N-terminal propeptides in the extracellular matrix are the cause of arthrochalasia EDS (aEDS). Autosomal recessive mutations in the gene encoding procollagen N-peptidase result in dermatosparaxis EDS (dEDS).
- (e) Fibril formation: dominant-negative pathogenic variants in COL5A1 and COL5A2 can prevent collagen molecules from assembling into heterotopic fibrils and cause approximately 30% of classical EDS (cEDS).
- (f) Interaction with extracellular matrix proteins: if the tissue-specific arrangement of collagen fibrils and the interaction with extracellular matrix proteins such as tenascin-X is disturbed, EDS with tenascin-X deficiency or classical-like EDS (clEDS) may result.
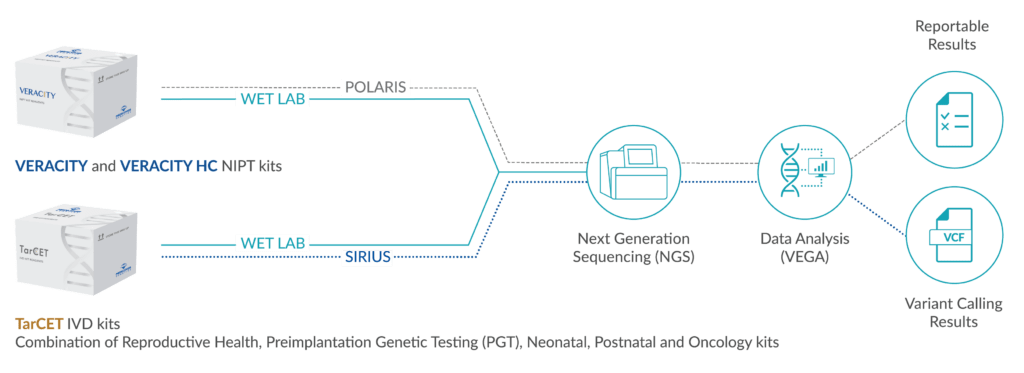
Since the clinical differentiation of individual EDS subtypes is often difficult and there may be overlap with other connective tissue diseases such as cutis laxa, genetic diagnosis using NGS may help in classification.
Ehlers-Danlos syndrome
29 genes: ADAMTS2, AEBP1, B3GALT6, B4GALT7, C1R, C1S, CHST14, COL12A1, COL1A1, COL1A2, COL3A1, COL5A1, COL5A2, COL6A1, COL6A2, COL6A3, DSE, EMILIN1, FKBP14, FLNA, PHYKPL, PIEZO2, PLOD1, PLOD3, PRDM5, SLC2A10, SLC39A13, TNXB, ZNF469
5 genes: COL1A1, COL1A2, COL3A1, COL5A1, COL5A2
11 genes: ADAMTS2, AEBP1, B3GALT6, B4GALT7, CHST14, COL1A2, DSE, FKBP14, PLOD1, SLC39A13, TNXB
EDS, rare forms, differential diagnoses
14 genes: C1R, C1S, COL12A1, COL6A1, COL6A2, COL6A3, EMILIN1, FLNA, PHYKPL, PIEZO2, PLOD3, PRDM5, SLC2A10, ZNF469
References
Cortini et al. 2019, Arch Dermatol Res 311:265 / Mayer in Luttkus (Hrsg.) 2018: Das Ehlers-Danlos-Syndrom, Kap. 3, 2. Auflage, De Gruyter / Weerakkody et al. 2018, Genet Med 18:1119 / Malfait et al. 2017, Am J Med Genet C 175:8 / Bowen et al. 2017, Am J Med Genet C 175:27 / Byers et al. 2017, Am J Med Genet C 175:40-47 / Brady et al. 2017, Am J Med Genet C 175:70 / Mayer et al. 2013, Eur J Hum Genet 21, update 2012 / Mayer in eLS 2012, John Wiley & Sons Ltd: Chichester http://www.els.net/ [DOI: 10.1002/9780470015902.a0024295] / Beighton et al. 1998, Am J Med Genet 77:31 / Hausser et al. 1994, Hum Genet 93:394