OVERVIEW
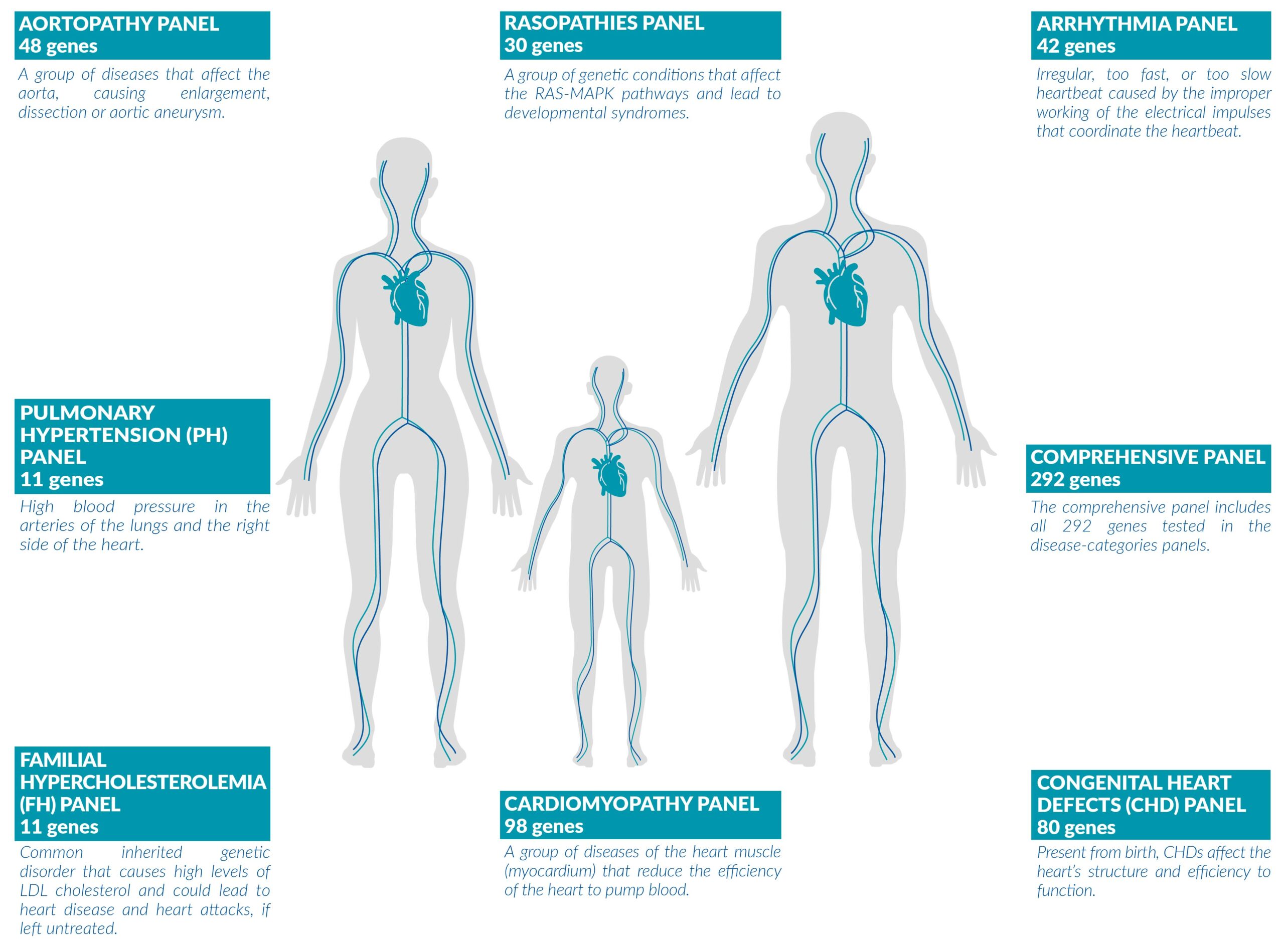
WHAT IS VENTRILIA?Ventrilia is a genetic test that analyzes up to 292 genes to detect mutations that can cause multiple cardiovascular conditions with complex phenotypes. It can be used to identify genetic mutations associated with cardiovascular diseases in:
• symptomatic individuals with a clinically diagnosed cardiovascular condition
• presymptomatic individuals with a clinically diagnosed cardiovascular condition
• asymptomatic individuals that belong in high-risk groups
Cardiovascular diseases are the leading cause of illness and death worldwide, responsible for 31% of all global deaths*. Previously, assessment of the disease risk was based on the lifestyle of an individual. The role of genetic testing is now becoming increasingly important, as overlapping symptoms make it very challenging to clearly identify the underlying cardiovascular condition. Genetic testing can detect the underlying condition, provide a prognosis and identify at-risk family members, who might be predisposed to the same cardiovascular disease. Identification of a genetic mutation that causes a disease, can lead to an improved prognosis as well as effective clinical management and treatment for many cardiovascular conditions†.
*World Health Organization (Cardiovascular Diseases fact sheets and World Heart Federation (2017)
†Clinical Appropriateness Guidelines – Genetic Testing for Hereditary Cardiac Disease – developed by Informed Medical Decisions, Inc.(2019)

OUR TESTS
Ventrilia tests detect clinically actionable genetic alterations, including Single Nucleotide Variants (SNVs), Insertions and Deletions (INDELs), Copy Number Amplifications (CNAs), and Rearrangements, that are associated with cardiovascular diseases.
Buccal swab collection device, which is provided in the Ventrilia kit
2-4 weeks from sample receipt in our laboratory
gDNA is extracted using a standardized methodology and subjected to mechanical fragmentation prior to DNA library preparation. DNA enrichment for the genomic regions of interest is carried out using a solution-based hybridization method, followed by next generation sequencing (NGS). Single nucleotide variants, small insertions and deletions (≤30bp), and copy number variations (CNVs) are reported. All positive CNVs are confirmed using an orthogonal method. The test aims to detect all coding exons, of MANE and/or Canonical transcripts, and 10bp of adjacent intronic sequence.
Exceptions may include: regions containing repeats, sequences of high homology such as segmental duplications and pseudogenes, as well as regions of extreme GC-content.
Variants are classified according to the criteria set by the American College of Medical Genetics and Genomics. Classification and interpretation of variants is performed using the Varsome Clinical platform, and is according to the published knowledge at the time of testing.
Next generation sequencing (Illumina)
Proprietary Target Capture Enrichment Technology (Click here to see our Publications)
GATK and Vardict variant callers. Targeted CNV calling using a proprietary bioinformatics pipeline utilizing a circular binary segmentation algorithm
Varsome Clinical by Saphetor
hg19, NCBI GRCh37
>30 (precision 99,9%)
Minimum percentage of bases with RD greater than 20X = 97%
Sensitivity: 100% (79.4%-100%)
Specificity: 100% (99.8%-100%)
Richards et al. 2015, Genet Med 17:405; ClinGen Sequence Variant Interpretation Recommendation for PM2-Version 1.0; Tavtigian SV et al. 2020, Hum Mutat 41(10); Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines; Pejaver V et al. 2022, bioRxiv;
ALoFT, BayesDel, DEOGEN2, Eigen, Eigen-PC, FATHMM, FATHMM-XF, FATHMM-MKL, fitCons, LIST-S2, LRT, M-CAP, MetaLR, MetaRNN, MetaSVM, MPC, MutationAssessor, MutationTaster, MutPred, MVP, Polyphen-2, PrimateAI, PROVEAN, REVEL, SIFT, SIFT4G, dbscSNV
More than 100 datasets including ClinVar, gnomAD, ExAC, ClinGen, DECIPHER, etc
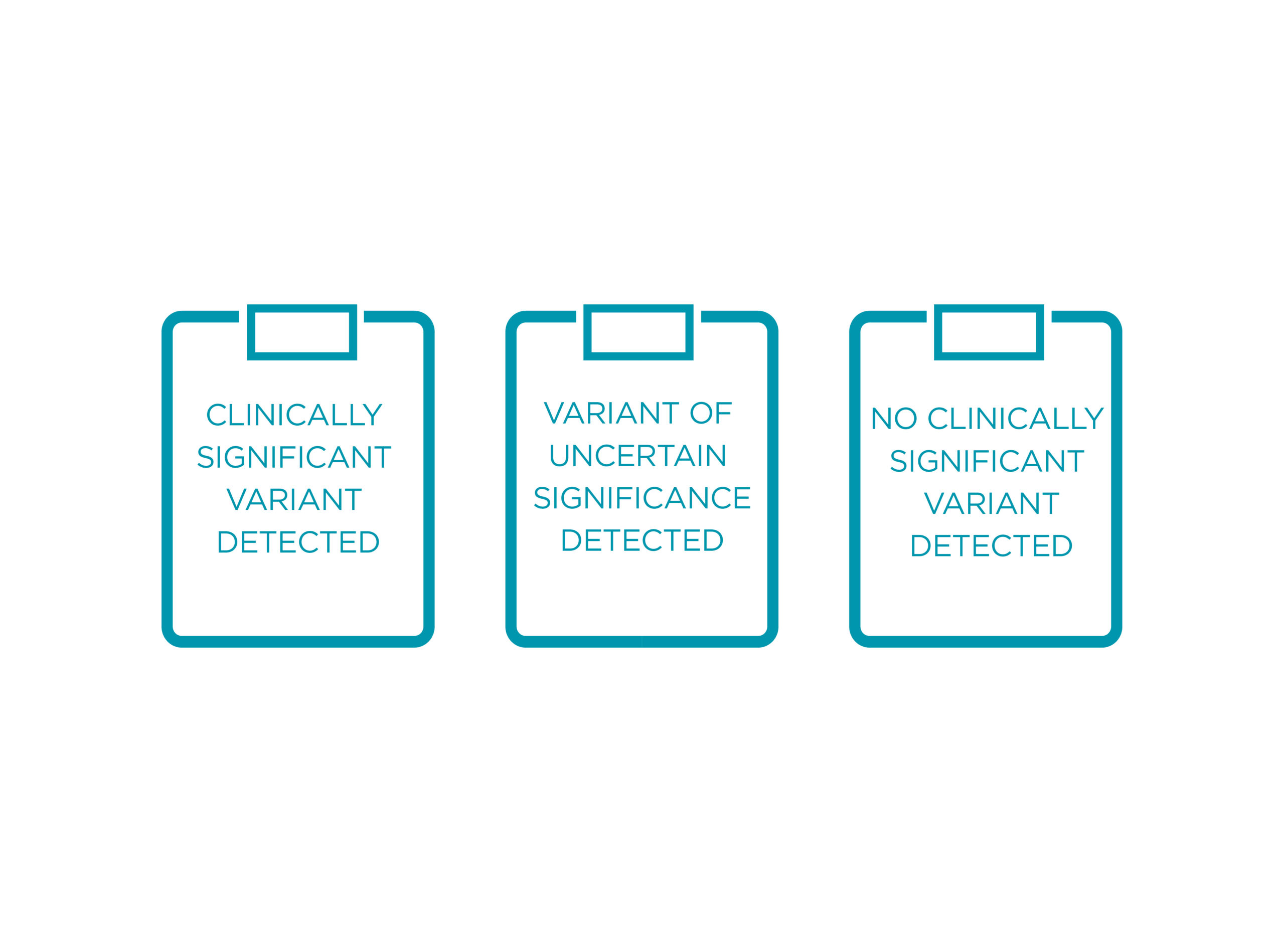
POSSIBLE OUTCOMES OF THE TEST
The Ventrilia report will have information on the following:
• Results on genes tested
• Thorough interpretation and clinical significance of mutations detected
Ventrilia reports on pathogenic and likely pathogenic variants, as well as variants of uncertain significance
• Carrier status will not be reported for recessive conditions
• VUS will only be reported in cases of potential pathogenicity