OVERVIEW
Medicover Genetics TarCET IVD kits offer a wide range of reliable and easy to implement genetic testing solutions, providing accurate, fast and cost efficient detection of genetic variants.
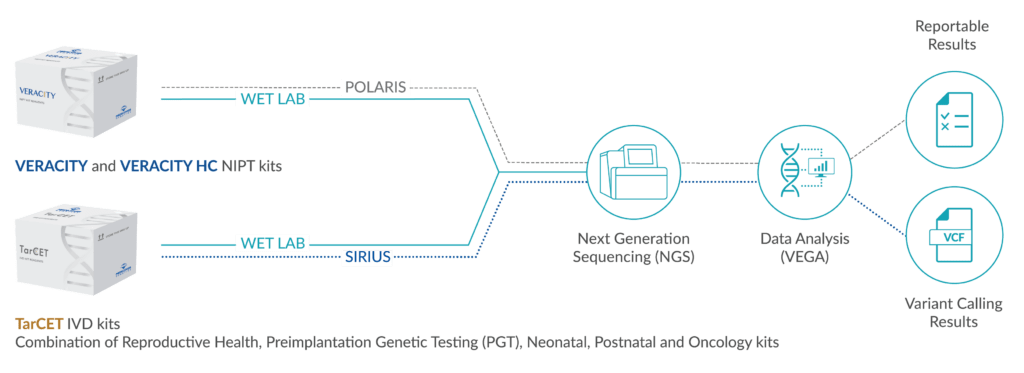
TarCET IVD kits are NGS-based, CE-marked, multi-gene kits that include reagents for a universal library preparation and target capture enrichment workflow covering multiple disciplines. All TarCET IVD kits assays can be combined into a consolidated workflow, by integrating multiple assays into the same sequencing run. With the unique capacity to run multidiscipline genetic tests in a single sequencing run, Medicover Genetics’ TarCET IVD kits ensure accurate and fast detection of genetic variants with cost and operational efficiencies for laboratories of any throughput capacities.
TarCET IVD Kits are CE marked library preparation and enrichment products, produced under strict quality-controlled manufacturing processes (ISO 13485:2016 and ISO 9001:2015), ensuring the highest quality, and intended to be used for identification of genetic variants associated with diseases.


FEATURES & SPECIFICATIONS
● Universal protocol for all assays
● Streamlined simple workflow
● Universal sample input requirements
● Automation compatibility
● Scalable for any throughput laboratory
● Specimen type: Buccal swab (except PGT; blastomeres or blastocysts biopsies)
● Mean coverage uniformity ≥ 20x: >97%
● Proprietary bioinformatics
● Unique dual-index combinations compatible with all TarCET IVD kits

ASSAY MENU
Medicover Genetics TarCET IVD kits offer a unique and efficient solution to any laboratory throughput to perform multi-disciplinary genetic tests in-house. By combining different assays in one single sequencing run, laboratories can improve turnaround time and reduce operational costs while ensuring high-quality results. Our comprehensive testing approach is ideal for laboratories looking to provide a wide range of high-fidelity genetic tests to their healthcare associates and patients.
TarCET IVD kits cover the following disciplines:
| Kit | Description |
| Carrier Screening Core (see more) | Analyzes 19 genes in individuals with unknown carrier status. |
| Carrier Screening Comprehensive (see more) | Analyzes 228 genes in individuals with unknown carrier status. |
| PGT (see more) | Detection of whole chromosome aneuploidies, structural rearrangements and segmental aneuploidies down to 10Mb, selected male polyploidies, and mosaicism higher than 50%. |
| Hereditary Cancer (see more) | Analyzes 62 genes and covers 24 cancer predisposing syndromes associated with hereditary cancer. |
| Infertility (see more) | Analyzes 54 genes in the female infertility panel and 39 genes in the male infertility panel. Structural and numerical abnormalities on sex chromosomes are included in both panels. |
| Neonatal (see more) | Analyzes 140 genes in symptomatic and pre-symptomatic infants. |
| Cardiac Comprehensive (see more) | Analyzes 292 genes and covers major inherited cardiovascular disorders. |
| Cardiomyopathy (see more) | Analyzes 98 genes and covers cardiomyopathy-related inherited cardiovascular disorders. |
| Arrhythmia (see more) | Analyzes 42 genes and covers arrhythmia-related inherited cardiovascular disorders. |
| Aortopathy (see more) | Analyzes 48 genes and covers aortopathy-related inherited cardiovascular disorders. |
| Congenital Heart Defects (see more) | Analyzes 80 genes and covers inherited congenital heart defects. |
| FH, PH and RAS (see more) | Analyzes 11, 11, and 30 genes and covers Familial Hypercholesterolemia (FH), Pulmonary Hypertension (PH) and RASopathy (RAS) related disorders, respectively. |
| Metabolic (see more) | Analyzes 223 genes and covers major classes of inherited metabolic disorders. |
| UltraVerse Index Oligos Type A/B/C/D | UltraVerse Index Oligos include unique dual index oligos compatible with all TarCET IVD Kits. |
Each TarCET IVD kit contains reagents for 16 reactions with multiplexing capabilities up to 384 (4 x 96-index kits). Each UltraVerse Index Oligos kit includes 96 reactions.
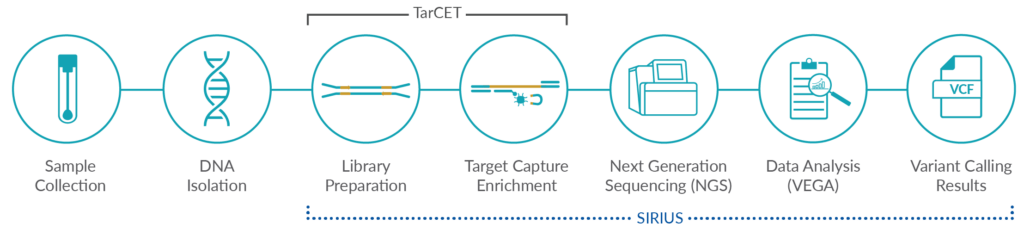
SIRIUS
SIRIUS is a genetic data management web application that enables users to manage information generated by NGS analysis when samples are being processed using TarCET IVD kits workflow. Additionally, SIRIUS facilitates the creation, calculation, and modification of sample batches corresponding to sequencing runs and interfaces with the Medicover Genetics analysis engine VEGA, to provide information on the analysis data in visual graphs and tabular formats.


VEGA
VEGA is a bioinformatics analysis software for samples that have been processed using TarCET IVD kits. VEGA analyzes NGS data generated by TarCET IVD workflow. The analysis detects single nucleotide variants, short insertions and/or deletions and copy number alterations. PGT analysis detects aneuploidies and structural rearrangements.
| Product Name | Catalog Number |
| TarCET Metabolic Kit | ET101-00-2016 |
| TarCET PGT Kit | ET102-00-2016 |
| TarCET Aortopathy Kit | ET103-00-2016 |
| TarCET Arrhythmia Kit | ET104-00-2016 |
| TarCET Cardiomyopathy Kit | ET105-00-2016 |
| TarCET Congenital Heart Defects Kit | ET106-00-2016 |
| TarCET FH, PH and RAS Kit | ET107-00-2016 |
| TarCET Cardiac Comprehensive Kit | ET110-00-2016 |
| TarCET Hereditary Cancer Kit | ET111-00-2016 |
| TarCET Infertility Kit | ET112-00-2016 |
| TarCET Neonatal Kit | ET113-00-2016 |
| TarCET Carrier Screening Core Kit | ET114-00-2016 |
| TarCET Carrier Screening Comprehensive Kit | ET115-00-2016 |
| UltraVerse Index Oligos Type A | EU100-00-100A |
| UltraVerse Index Oligos Type B | EU100-00-100B |
| UltraVerse Index Oligos Type C | EU100-00-100C |
| UltraVerse Index Oligos Type D | EU100-00-100D |
How to order: customersupport.genetics@medicover.com
SUPPORT
Medicover Genetics offers on-going support to all TarCET IVD kits customers through dedicated technical support channels. Members of our team are always available to offer technical support on troubleshooting, quality and performance monitoring. Upon request, we provide customer trainings on workflow, protocol, and data analysis in our certified facilities.
For technical support inquiries: ivdsupport.genetics@medicover.com

DOCUMENTATION
TarCET IVD kits Instructions for use
TarCET PGT kit Instructions for use
UltraVerse Instructions for use
Declaration of conformity for TarCET PGT KIT
Declaration of conformity TarCET IVD KIT
Registration certificate for TarCET IVD KIT
Registration certificate for TarCET PGT KIT