OVERVIEW
WHAT IS ForeSENTIA?ForeSENTIA is a tumor profiling genetic test that can identify a spectrum of genetic alterations and genomic biomarkers, such as Microsatellite Instability (MSI) and Tumor Mutational Burden (TMB).
The information provided by ForeSENTIA can offer guidance on treatment decisions, prognostic value, and improved clinical management through precision medicine and targeted therapies.
● Option to test for clinically actionable genetic alterations, and immunotherapy biomarkers MSI and TMB in a single test
● Testing of thoroughly selected genetic alterations and biomarkers:
• Genes recommended by NCCN guidelines for solid tumors
• Genes that currently serve as selection criteria in active clinical trials
● Tumor-agnostic biomarkers
● Guidance on available targeted therapies, including immunotherapies. Tailored therapies can reduce the risk of ineffective therapy and adverse side effects, and avoid a ‘one-size-fits-all’ approach.

WHO IS ForeSENTIA FOR?
Patients diagnosed with a
solid type of tumor and
require genetic test analysis for targeted therapy
Patients with treatment resistance
who require alternative treatment options,
including immunotherapy
Patients diagnosed with a
solid type of tumor and want to increase
the possibilities of identifying treatment options
by testing for immunotherapy biomarkers MSI and TMB
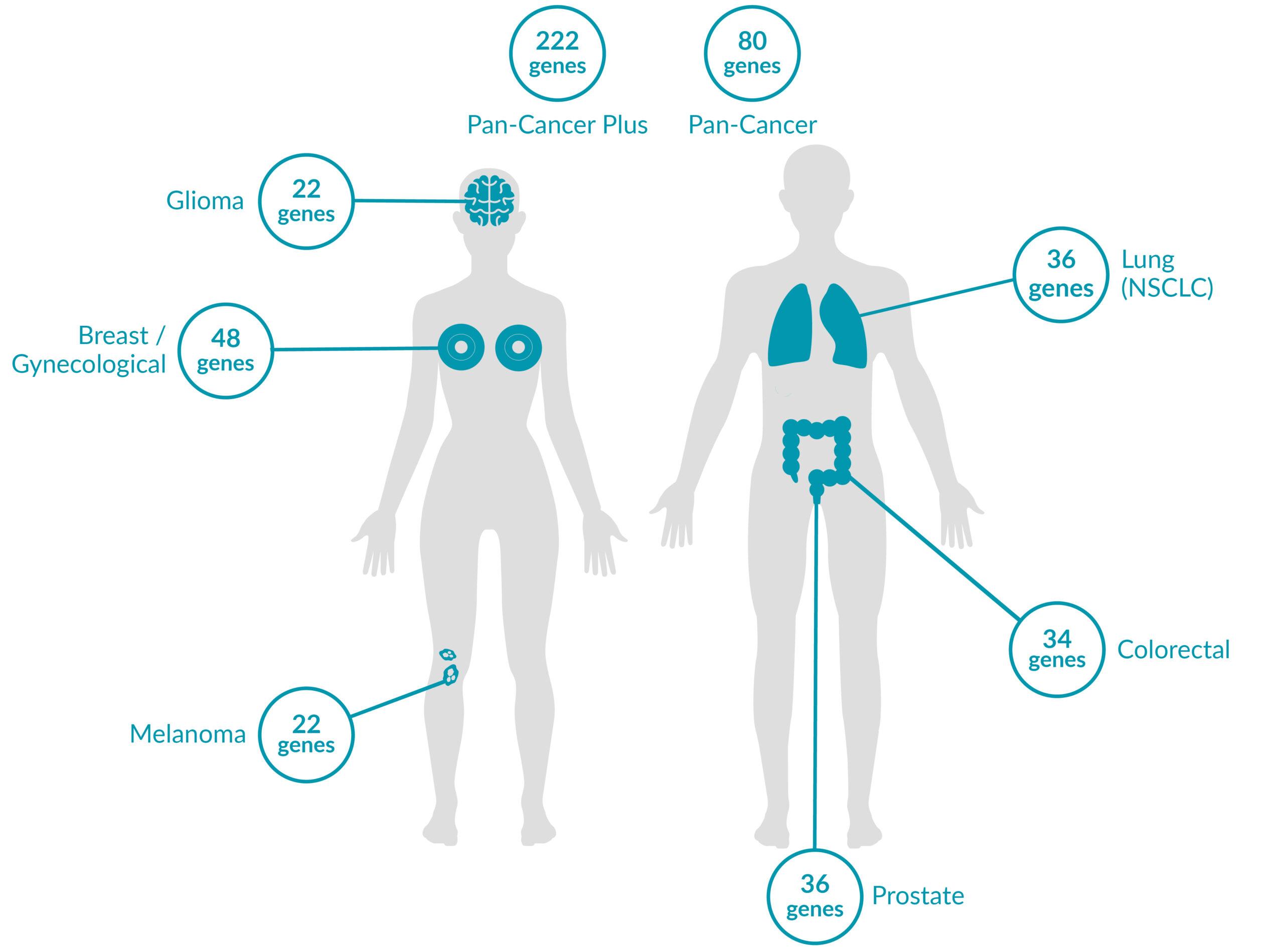
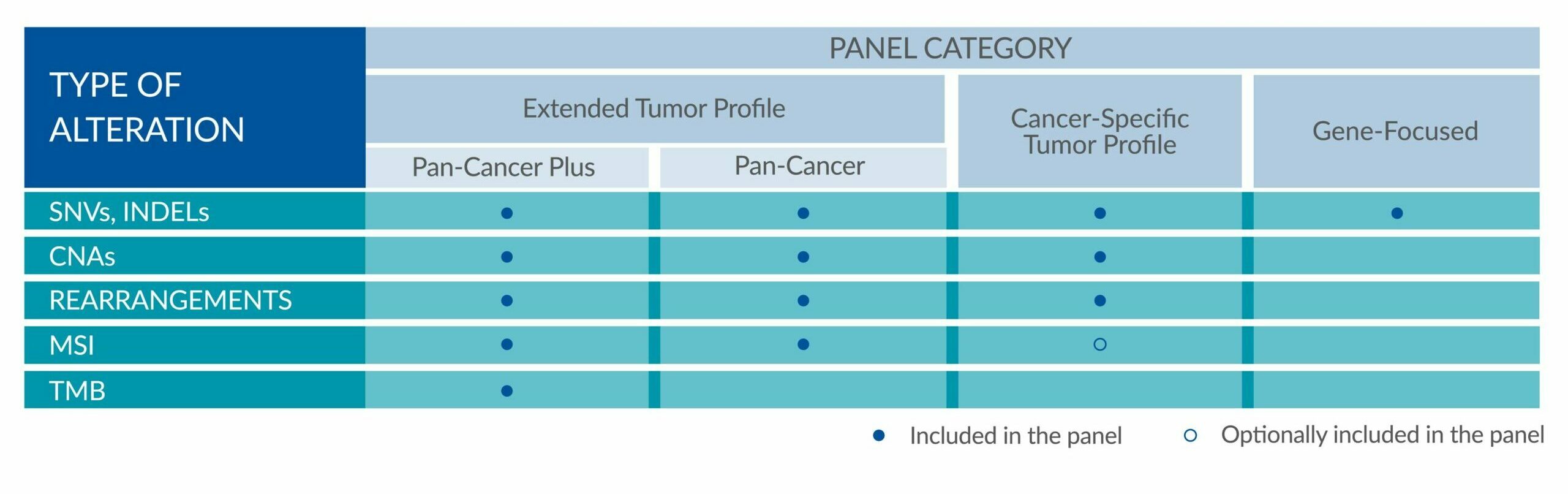
Extended tumor profile panels target genetic alterations in an extended number of genes, and include testing for MSI via Next Generation Sequencing (NGS).
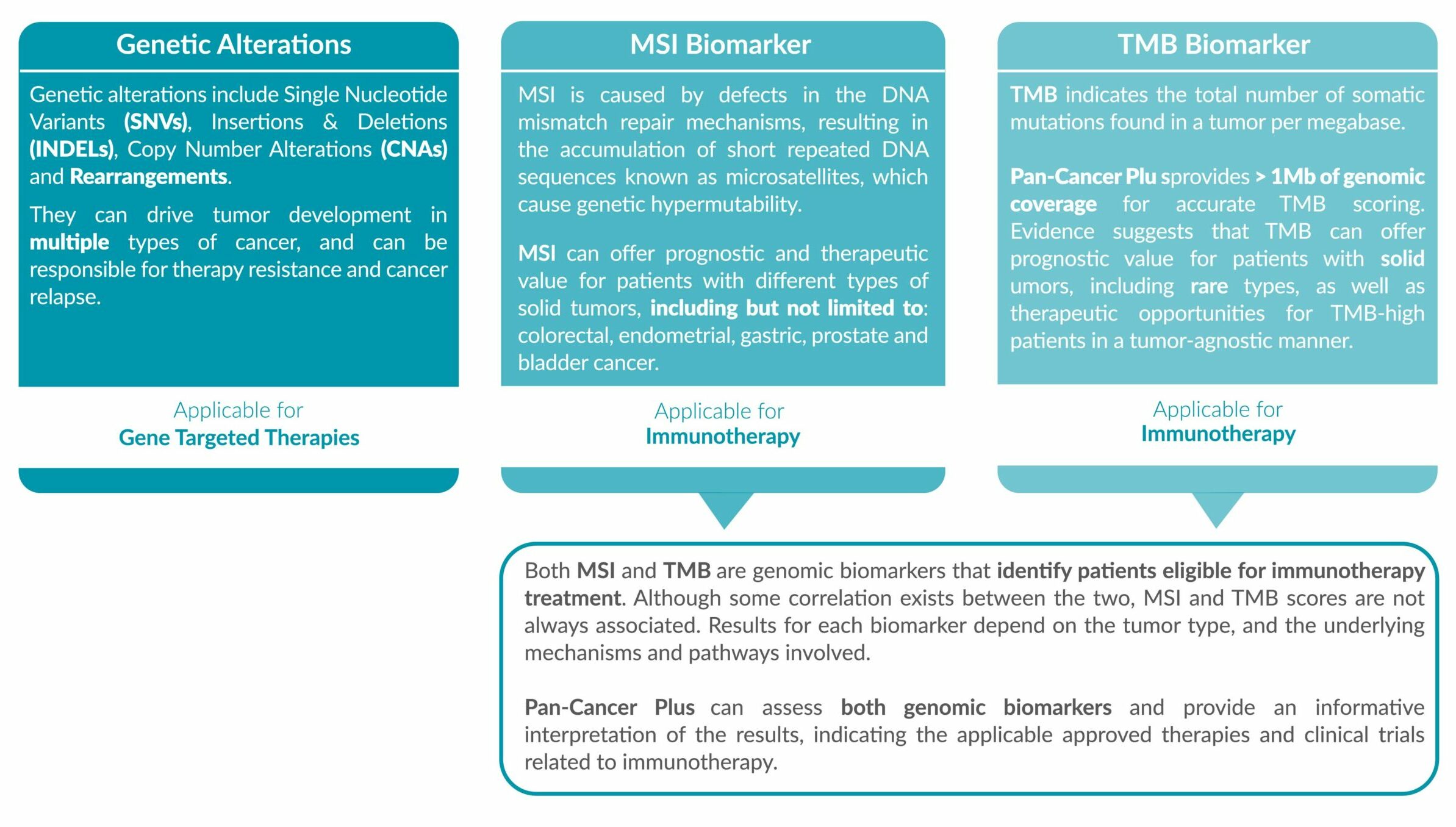
Genetic alterations tested include single nucleotide variants (SNVs), insertions and deletions (INDELs), copy number alterations (CNAs) and rearrangements.
ForeSENTIA Pan-Cancer Plus tests for all exonic regions* in an extended number of genes and includes immunotherapy biomarkers MSI and TMB assessment via NGS
*Exceptions on regions containing repeats, sequences of high homology (pseudogenes and segmental duplications) or high GC-content.
Download here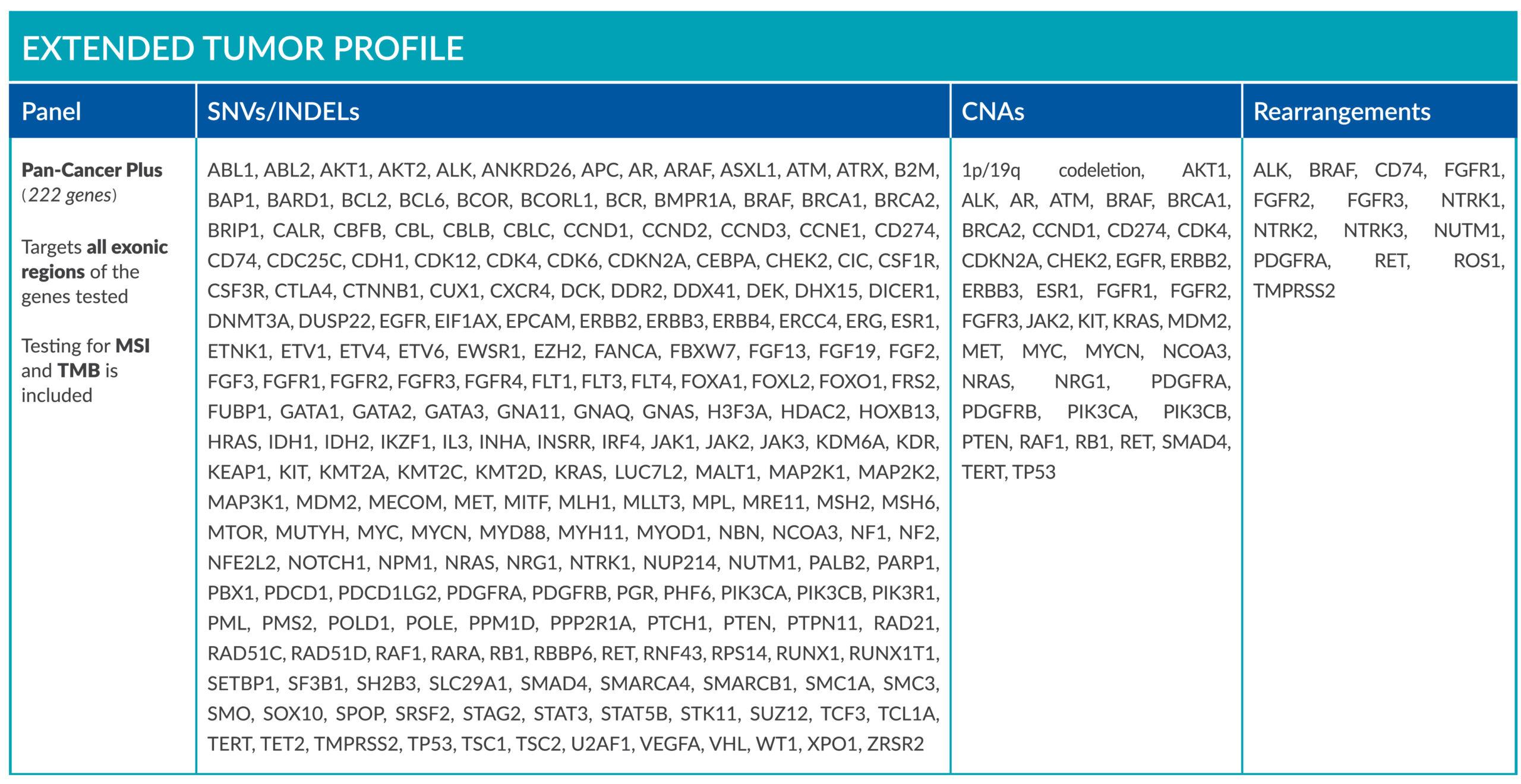
ForeSENTIA Pan-Cancer tests for an extended number of genes and includes immunotherapy biomarker MSI assessment via NGS.
Download here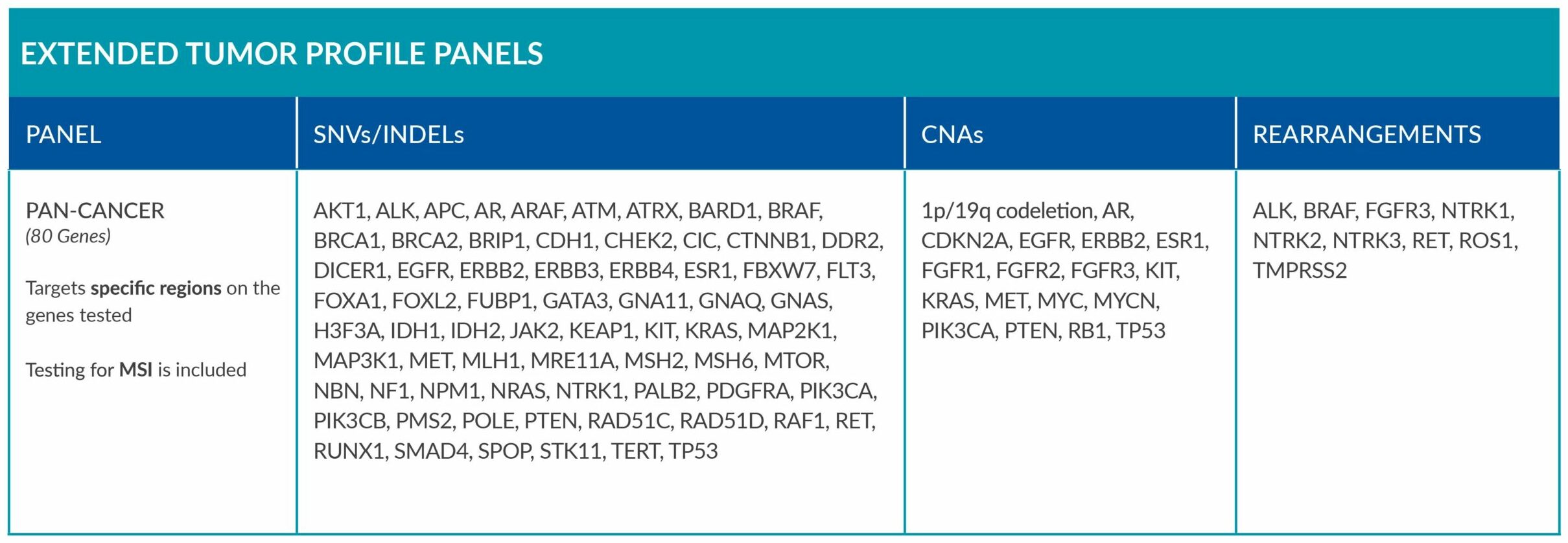
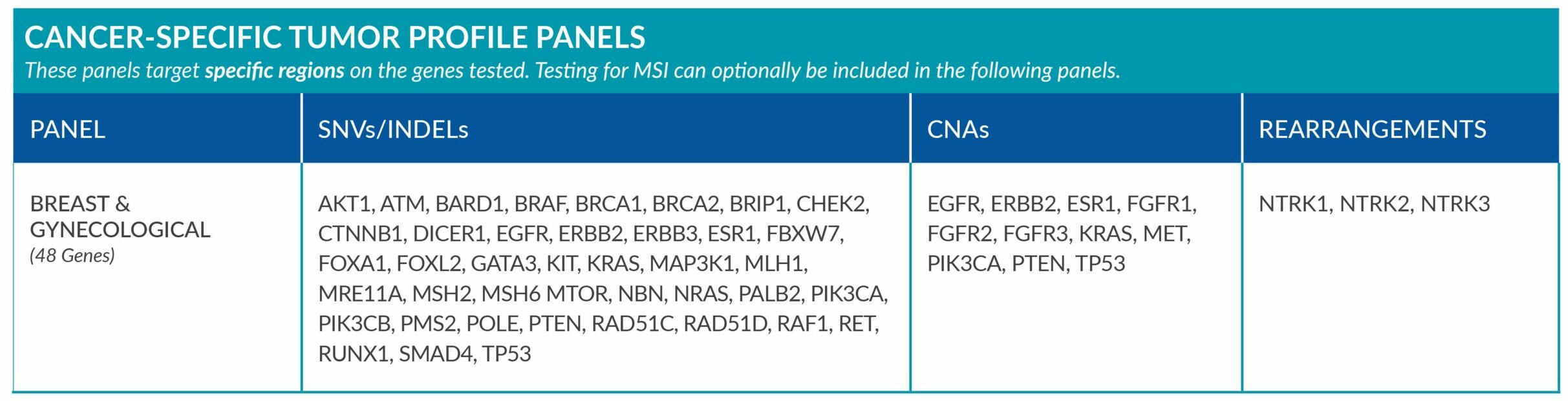
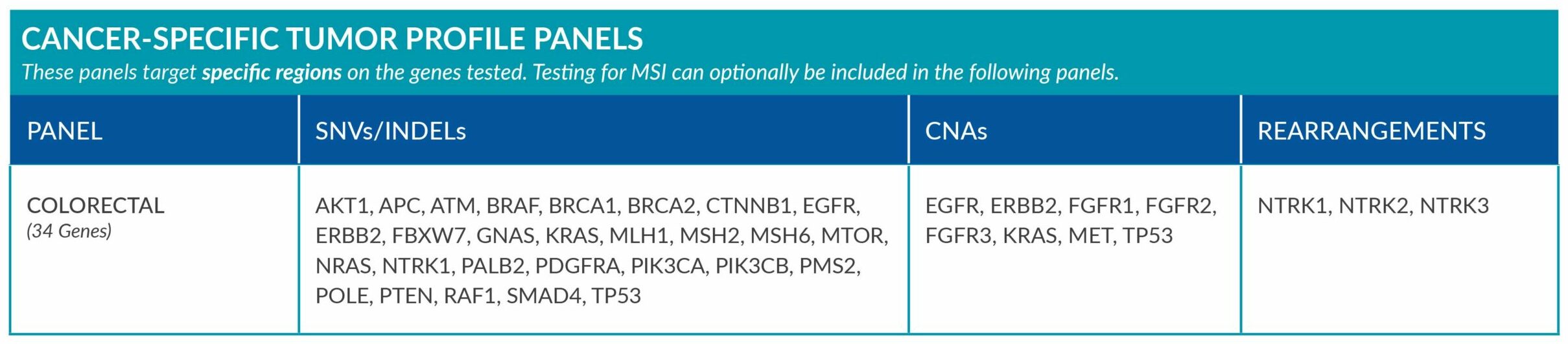
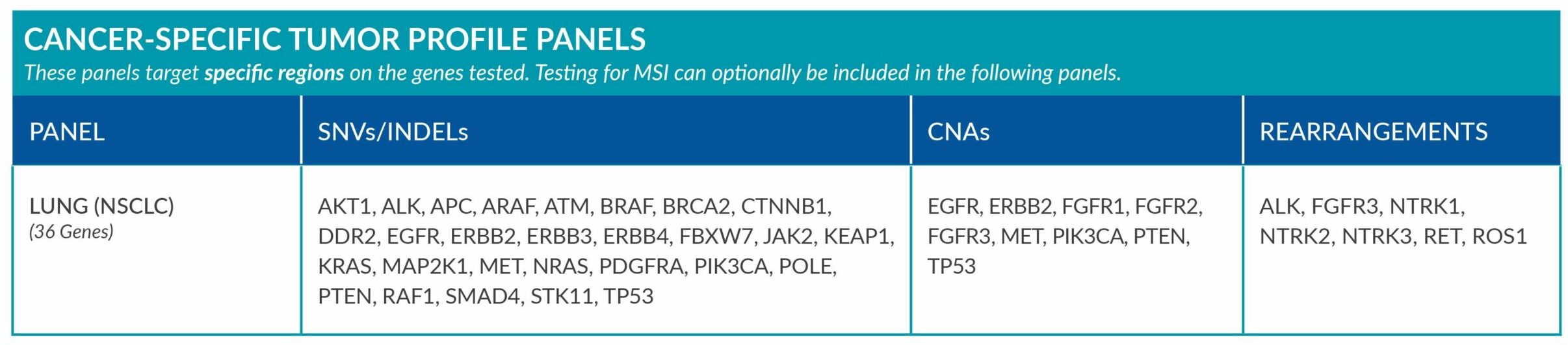
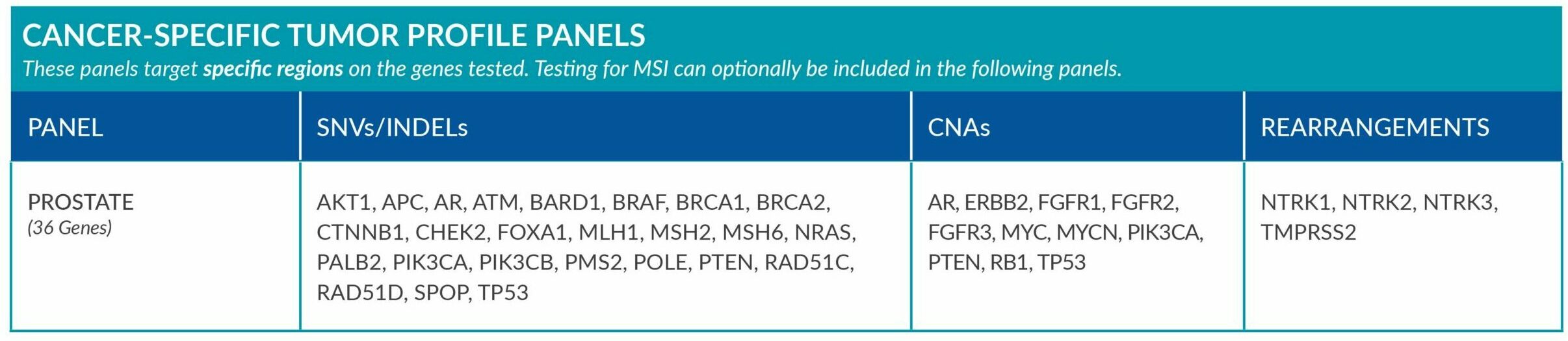
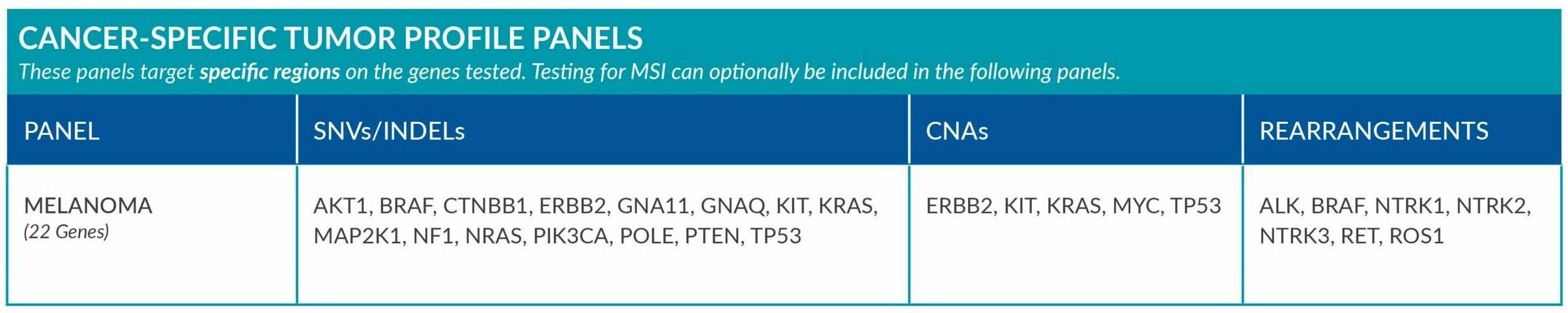
Cancer-Specific tumor profile panels target specific genetic alterations in several genes, and can optionally include MSI assessment via NGS. Genetic alterations tested include single nucleotide variants (SNVs), insertions and deletions (INDELs), copy number alterations (CNAs) and rearrangements.
Gene-Focused panels target genetic alterations in one or two genes. Genetic alterations tested include single nucleotide variants (SNVs), insertions and deletions (INDELs).
GENETIC ALTERATIONS AND BIOMARKERS TESTED PER PANEL
TESTING OF GENETIC ALTERATIONS AND GENOMIC BIOMAKERS
POSSIBLE OUTCOMES OF THE TEST
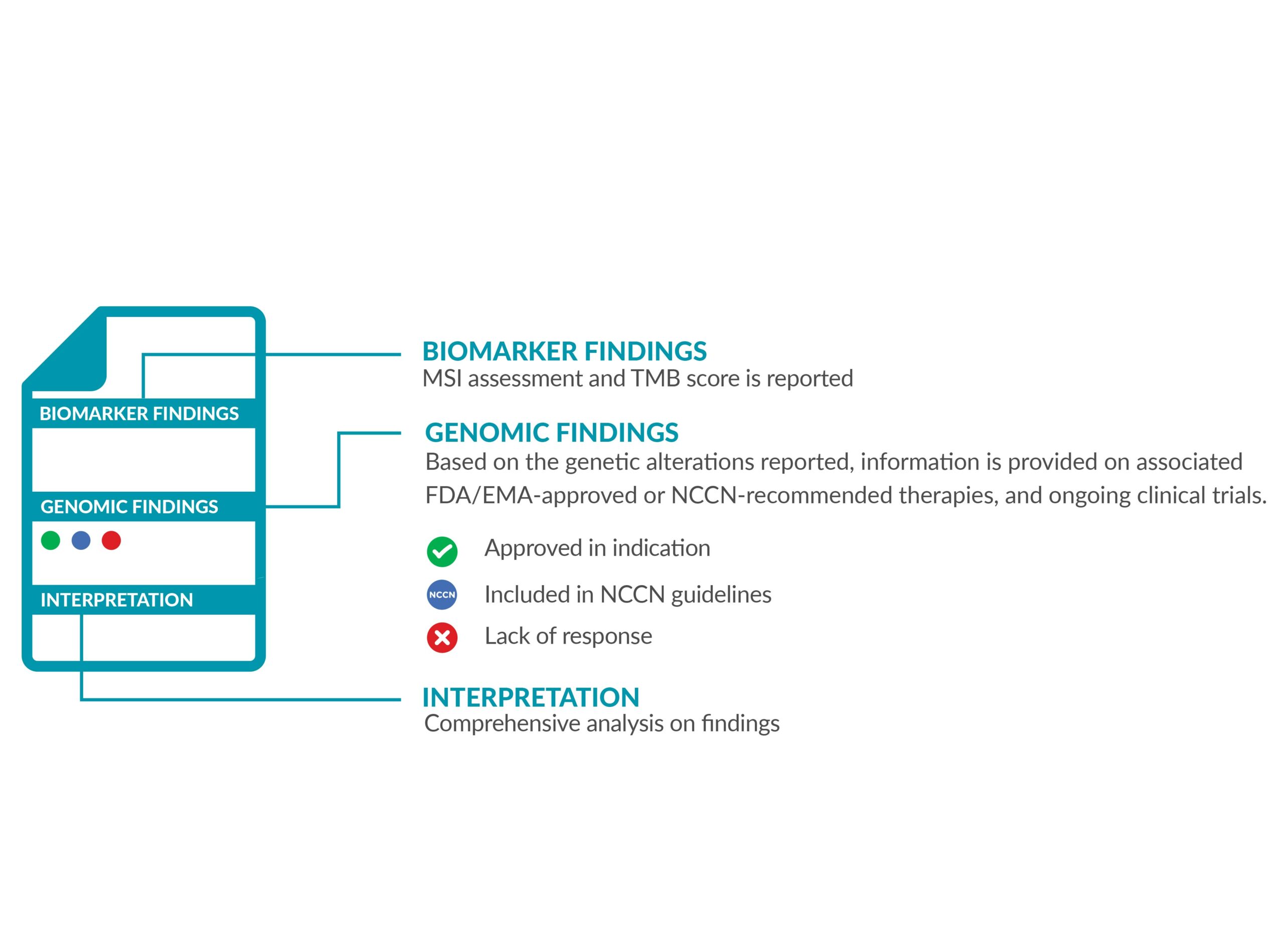
The ForeSENTIA report will have information on the following:
• IMMUNOTHERAPY BIOMARKERS
MSI status and TMB score is reported, accompanied with the FDA/EMA-approved drugs which are eligible for the biomarker detected.
• GENOMIC FINDINGS
All the genetic variants (alterations) identified are reported. The results will also indicate which approved FDA/EMA targeted therapies are suitable as treatment options. Depending on the results identified, the report will also provide information on the drugs the patient will not benefit from.
Variants of strong and of potential clinical significance as well as variants of uncertain significance (VUS) are also reported.
• CLINICAL TRIALS
Information on clinical trials which are relevant to the genetic alterations identified.
• INTERPRETATION
The biological and therapeutical classification of results are reported in detail according to NCCN guidelines